Second-Tier DNA Confirmation of Newborn Screening Results
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Inherited Metabolic Disease
Inherited metabolic disease Dr Neil W Hopper SRH Areas for discussion • Introduction to IEMs • Presentation • Initial treatment and investigation of IEMs • Hypoglycaemia • Hyperammonaemia • Other presentations • Management of intercurrent illness • Chronic management Inherited Metabolic Diseases • Result from a block to an essential pathway in the body's metabolism. • Huge number of conditions • All rare – very rare (except for one – 1:500) • Presentation can be non-specific so index of suspicion important • Mostly AR inheritance – ask about consanguinity Incidence (W. Midlands) • Amino acid disorders (excluding phenylketonuria) — 18.7 per 100,000 • Phenylketonuria — 8.1 per 100,000 • Organic acidemias — 12.6 per 100,000 • Urea cycle diseases — 4.5 per 100,000 • Glycogen storage diseases — 6.8 per 100,000 • Lysosomal storage diseases — 19.3 per 100,000 • Peroxisomal disorders — 7.4 per 100,000 • Mitochondrial diseases — 20.3 per 100,000 Pathophysiological classification • Disorders that result in toxic accumulation – Disorders of protein metabolism (eg, amino acidopathies, organic acidopathies, urea cycle defects) – Disorders of carbohydrate intolerance – Lysosomal storage disorders • Disorders of energy production, utilization – Fatty acid oxidation defects – Disorders of carbohydrate utilization, production (ie, glycogen storage disorders, disorders of gluconeogenesis and glycogenolysis) – Mitochondrial disorders – Peroxisomal disorders IMD presentations • ? IMD presentations • Screening – MCAD, PKU • Progressive unexplained neonatal -

Amino Acid Disorders
471 Review Article on Inborn Errors of Metabolism Page 1 of 10 Amino acid disorders Ermal Aliu1, Shibani Kanungo2, Georgianne L. Arnold1 1Children’s Hospital of Pittsburgh, University of Pittsburgh School of Medicine, Pittsburgh, PA, USA; 2Western Michigan University Homer Stryker MD School of Medicine, Kalamazoo, MI, USA Contributions: (I) Conception and design: S Kanungo, GL Arnold; (II) Administrative support: S Kanungo; (III) Provision of study materials or patients: None; (IV) Collection and assembly of data: E Aliu, GL Arnold; (V) Data analysis and interpretation: None; (VI) Manuscript writing: All authors; (VII) Final approval of manuscript: All authors. Correspondence to: Georgianne L. Arnold, MD. UPMC Children’s Hospital of Pittsburgh, 4401 Penn Avenue, Suite 1200, Pittsburgh, PA 15224, USA. Email: [email protected]. Abstract: Amino acids serve as key building blocks and as an energy source for cell repair, survival, regeneration and growth. Each amino acid has an amino group, a carboxylic acid, and a unique carbon structure. Human utilize 21 different amino acids; most of these can be synthesized endogenously, but 9 are “essential” in that they must be ingested in the diet. In addition to their role as building blocks of protein, amino acids are key energy source (ketogenic, glucogenic or both), are building blocks of Kreb’s (aka TCA) cycle intermediates and other metabolites, and recycled as needed. A metabolic defect in the metabolism of tyrosine (homogentisic acid oxidase deficiency) historically defined Archibald Garrod as key architect in linking biochemistry, genetics and medicine and creation of the term ‘Inborn Error of Metabolism’ (IEM). The key concept of a single gene defect leading to a single enzyme dysfunction, leading to “intoxication” with a precursor in the metabolic pathway was vital to linking genetics and metabolic disorders and developing screening and treatment approaches as described in other chapters in this issue. -

Centometabolic® COMBINING GENETIC and BIOCHEMICAL TESTING for the FAST and COMPREHENSIVE DIAGNOSTIC of METABOLIC DISORDERS
CentoMetabolic® COMBINING GENETIC AND BIOCHEMICAL TESTING FOR THE FAST AND COMPREHENSIVE DIAGNOSTIC OF METABOLIC DISORDERS GENE ASSOCIATED DISEASE(S) ABCA1 HDL deficiency, familial, 1; Tangier disease ABCB4 Cholestasis, progressive familial intrahepatic 3 ABCC2 Dubin-Johnson syndrome ABCD1 Adrenoleukodystrophy; Adrenomyeloneuropathy, adult ABCD4 Methylmalonic aciduria and homocystinuria, cblJ type ABCG5 Sitosterolemia 2 ABCG8 Sitosterolemia 1 ACAT1 Alpha-methylacetoacetic aciduria ADA Adenosine deaminase deficiency AGA Aspartylglucosaminuria AGL Glycogen storage disease IIIa; Glycogen storage disease IIIb AGPS Rhizomelic chondrodysplasia punctata, type 3 AGXT Hyperoxaluria, primary, type 1 ALAD Porphyria, acute hepatic ALAS2 Anemia, sideroblastic, 1; Protoporphyria, erythropoietic, X-linked ALDH4A1 Hyperprolinemia, type II ALDOA Glycogen storage disease XII ALDOB Fructose intolerance, hereditary ALG3 Congenital disorder of glycosylation, type Id ALPL Hypophosphatasia, adult; Hypophosphatasia, childhood; Hypophosphatasia, infantile; Odontohypophosphatasia ANTXR2 Hyaline fibromatosis syndrome APOA2 Hypercholesterolemia, familial, modifier of APOA5 Hyperchylomicronemia, late-onset APOB Hypercholesterolemia, familial, 2 APOC2 Hyperlipoproteinemia, type Ib APOE Sea-blue histiocyte disease; Hyperlipoproteinemia, type III ARG1 Argininemia ARSA Metachromatic leukodystrophy ARSB Mucopolysaccharidosis type VI (Maroteaux-Lamy) 1 GENE ASSOCIATED DISEASE(S) ASAH1 Farber lipogranulomatosis; Spinal muscular atrophy with progressive myoclonic epilepsy -

Diseases Catalogue
Diseases catalogue AA Disorders of amino acid metabolism OMIM Group of disorders affecting genes that codify proteins involved in the catabolism of amino acids or in the functional maintenance of the different coenzymes. AA Alkaptonuria: homogentisate dioxygenase deficiency 203500 AA Phenylketonuria: phenylalanine hydroxylase (PAH) 261600 AA Defects of tetrahydrobiopterine (BH 4) metabolism: AA 6-Piruvoyl-tetrahydropterin synthase deficiency (PTS) 261640 AA Dihydropteridine reductase deficiency (DHPR) 261630 AA Pterin-carbinolamine dehydratase 126090 AA GTP cyclohydrolase I deficiency (GCH1) (autosomal recessive) 233910 AA GTP cyclohydrolase I deficiency (GCH1) (autosomal dominant): Segawa syndrome 600225 AA Sepiapterin reductase deficiency (SPR) 182125 AA Defects of sulfur amino acid metabolism: AA N(5,10)-methylene-tetrahydrofolate reductase deficiency (MTHFR) 236250 AA Homocystinuria due to cystathionine beta-synthase deficiency (CBS) 236200 AA Methionine adenosyltransferase deficiency 250850 AA Methionine synthase deficiency (MTR, cblG) 250940 AA Methionine synthase reductase deficiency; (MTRR, CblE) 236270 AA Sulfite oxidase deficiency 272300 AA Molybdenum cofactor deficiency: combined deficiency of sulfite oxidase and xanthine oxidase 252150 AA S-adenosylhomocysteine hydrolase deficiency 180960 AA Cystathioninuria 219500 AA Hyperhomocysteinemia 603174 AA Defects of gamma-glutathione cycle: glutathione synthetase deficiency (5-oxo-prolinuria) 266130 AA Defects of histidine metabolism: Histidinemia 235800 AA Defects of lysine and -
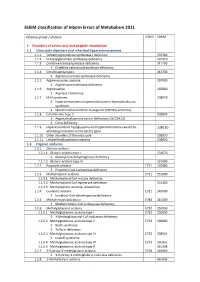
SSIEM Classification of Inborn Errors of Metabolism 2011
SSIEM classification of Inborn Errors of Metabolism 2011 Disease group / disease ICD10 OMIM 1. Disorders of amino acid and peptide metabolism 1.1. Urea cycle disorders and inherited hyperammonaemias 1.1.1. Carbamoylphosphate synthetase I deficiency 237300 1.1.2. N-Acetylglutamate synthetase deficiency 237310 1.1.3. Ornithine transcarbamylase deficiency 311250 S Ornithine carbamoyltransferase deficiency 1.1.4. Citrullinaemia type1 215700 S Argininosuccinate synthetase deficiency 1.1.5. Argininosuccinic aciduria 207900 S Argininosuccinate lyase deficiency 1.1.6. Argininaemia 207800 S Arginase I deficiency 1.1.7. HHH syndrome 238970 S Hyperammonaemia-hyperornithinaemia-homocitrullinuria syndrome S Mitochondrial ornithine transporter (ORNT1) deficiency 1.1.8. Citrullinemia Type 2 603859 S Aspartate glutamate carrier deficiency ( SLC25A13) S Citrin deficiency 1.1.9. Hyperinsulinemic hypoglycemia and hyperammonemia caused by 138130 activating mutations in the GLUD1 gene 1.1.10. Other disorders of the urea cycle 238970 1.1.11. Unspecified hyperammonaemia 238970 1.2. Organic acidurias 1.2.1. Glutaric aciduria 1.2.1.1. Glutaric aciduria type I 231670 S Glutaryl-CoA dehydrogenase deficiency 1.2.1.2. Glutaric aciduria type III 231690 1.2.2. Propionic aciduria E711 232000 S Propionyl-CoA-Carboxylase deficiency 1.2.3. Methylmalonic aciduria E711 251000 1.2.3.1. Methylmalonyl-CoA mutase deficiency 1.2.3.2. Methylmalonyl-CoA epimerase deficiency 251120 1.2.3.3. Methylmalonic aciduria, unspecified 1.2.4. Isovaleric aciduria E711 243500 S Isovaleryl-CoA dehydrogenase deficiency 1.2.5. Methylcrotonylglycinuria E744 210200 S Methylcrotonyl-CoA carboxylase deficiency 1.2.6. Methylglutaconic aciduria E712 250950 1.2.6.1. Methylglutaconic aciduria type I E712 250950 S 3-Methylglutaconyl-CoA hydratase deficiency 1.2.6.2. -

ERNDIM Diagnostic Proficiency Testing Is an Important Tool In
ERNDIM Diagnostic Proficiency Testing is an important Tool in Determining Quality of Laboratory Diagnosis in a wide Range of Inborn Errors of Metabolism Brian Fowler1; Jim Bonham2, Petr Chrastina3; Joanne Croft2; Viktor Kozich3; George Ruijter4; Christine Saban5 1.University Children’s Hospital, Basel/Zürich - Switzerland; 2. Children’s Hospital, Sheffield – United Kingdom; 3. Institute of Inherited Metabolic Diseases, Prague – Czech Republic; 4. Erasmus Medical Center, Rotterdam – Netherlands; 5. Service Maladies Héréditaires du Metabolisme, Bron-Lyon - France Background & Discussion Introduction Aims & Method Conclusions ERNDIM (www.erndim.org) aims to - 273 samples from 83 different conditions (some no IEM), sent - Overall performance is fairly good, emphasising that a improve the quality of diagnosis and to up to 105 laboratories, mainly European but also worldwide. definitive diagnosis is not always possible with urine alone. monitoring in patients with inborn errors - Organising centres - Czech Republic, France, Netherlands, - Evidence of improved performance repeat distributions of metabolism (IEM) through quality Switzerland, and United Kingdom, now sent centrally (CSCQ, - ERNDIM diagnostic proficiency testing valuable for: assurance programmes and educational Switzerland). • informing accreditation activities. Diagnostic proficiency testing - Six samples / year, one common to all centres, sent with • assessing individual laboratory performance schemes focus on the ability of clinical information. • identifying methodological and technical challenges laboratories to identify and interpret - Laboratories choose and perform tests (limited amount of • contributing to improved diagnostic approaches. abnormalities in natural urine samples urine) needed to reach a diagnosis using analysis of one or - Clinicians must be aware that laboratory testing for IEM can reflecting a wide range of IEMs. more of amino acids, organic acids, mucopolysaccharides, be difficult and results need to be viewed with caution. -

Liver Disorders in Inherited Metabolic Disorders
Steatosis/f Fibrosis/c Liver Name Hepatomegaly Elevated transaminases ALF Cholestasis atty liver irrhosis tumor Other Diagnostic markers Specific treatment Representative references (PMID or DOI) DISORDERS OF NITROGEN-CONTAINING COMPOUNDS Disorders of ammonia detoxification Protein restriction, ammonia scavengers N-acetylglutamate synthase deficiency X Ammonia (B), Urea (P), Amino acids (P) (carglumic acid), citrulline 28900784, 11131349 Protein restriction, ammonia Carbamoylphosphate synthetase I deficiency X X X X Ammonia (B), ASAT/ALAT (P), Urea (P), Amino acids (P) scavengers, citrulline, liver transplant 28900784 Ammonia (B), ASAT/ALAT (P), Urea (P), Amino acids (P), Protein restriction, ammonia 28900784, 22129577, 24485820, 21884343, Ornithine transcarbamylase deficiency X X X X X HCC Orotic acid (U) scavengers, citrulline, liver transplant 27070778, 24485820, 28887792 Ammonia (B), ASAT/ALAT (P), Urea (P), Amino acids (P), Protein restriction, ammonia Argininosuccinate synthetase deficiency X X X X HCC Orotic acid (U) scavengers, arginine, liver transplant 28900784, 29209134, 15334737 Ammonia (B), ASAT/ALAT (P), Urea (P), Amino acids (P, U), Protein restriction, ammonia Argininosuccinate lyase deficiency X X X X HCC Orotic acid (U) scavengers, arginine, liver transplant 28900784 Ammonia (B), ASAT/ALAT (P), Urea (P), Amino acids (P), Arginase deficiency X X X X X HCC Orotic acid (U) Protein restriction, ammonia scavengers 28900784, 22964440 Ammonia (B), ASAT/ALAT (P), Urea (P), Amino acids (P, U), Protein restriction, ammonia Mitochondrial -
Inborn Errors of Metabolism in Infancy
Inborn Errors of metabolism in Infancy 2002/01(制定) 2007/01(審) Clinical Manifestations Acute metabolic encephalopathy Metabolic acidosis Hyperammonemia Hypoglycemia Jaundice or hepatic dysfunction Dysmorphyism Acute metabolic encephalopathy Toxic effects of accumulating metabolites on the CNS Lethargy, poor feeding, sepsis-like(no risk factors), Seizures, abnormal muscle tone, cerebral edema, ICH apnea,tachypnea, recurrent vomiting, Basic laboratory studies CBC, DC Urinalysis Blood gases Serum electrolytes Blood glucose, lactate, CSF lactate Plasma ammonia Urine reducing substances Urine ketones if acidosis or hypoglycemia + Plasma and urine amino acids Urine organic acids Plasma lactate Urine metabolic screening Tandem mass (血片) Hyperammonemia Unexplained vomiting, lethargy, or other evidence of an encephalopahty Urea cycle defects, organic acidemias, transient hyperammonemia of the newborn, fatty acid oxidation defects Frequently > 1000μmol/L (v.s. < 500μmol/L in sepsis, herpes simplex infection, perinatal asphyxia) Metabolic acidosis Organic acidemias:Neutropenia, thrombocytopenia, elevated plasma lactate, hyperammonemia Defects in pyruvate metabolism, or in the respiratory chain Lactic acidosis , normal urine organic acid Plasma lactate/pyruvate ratio, < 25 defect in pyruvate dehydrogenase or in gluconeogenesis > 25 pyruvate carboxylase defeiciency, respiratory chain defect, mitochondrial myopathy Metabolic disease without metabolic acidosis or hyperammonemia nonketotic hyperglycinemia molybdenum co-factor deficiency Major inborn errors -

Amino Acids Profiling for the Diagnosis of Metabolic Disorders
Cleveland State University EngagedScholarship@CSU Chemistry Faculty Publications Chemistry Department 2019 Amino Acids Profiling for the Diagnosis of Metabolic Disorders Yana Sandlers Cleveland State University, [email protected] Follow this and additional works at: https://engagedscholarship.csuohio.edu/scichem_facpub Part of the Biochemistry Commons How does access to this work benefit ou?y Let us know! Recommended Citation Sandlers, Yana, "Amino Acids Profiling for the Diagnosis of Metabolic Disorders" (2019). Chemistry Faculty Publications. 552. https://engagedscholarship.csuohio.edu/scichem_facpub/552 This Contribution to Books is brought to you for free and open access by the Chemistry Department at EngagedScholarship@CSU. It has been accepted for inclusion in Chemistry Faculty Publications by an authorized administrator of EngagedScholarship@CSU. For more information, please contact [email protected]. We are IntechOpen, the world’s leading publisher of Open Access books Built by scientists, for scientists 5,000 126,000 145M Open access books available International authors and editors Downloads Our authors are among the 154 TOP 1% 12.2% Countries delivered to most cited scientists Contributors from top 500 universities Selection of our books indexed in the Book Citation Index in Web of Science™ Core Collection (BKCI) Interested in publishing with us? Contact [email protected] Numbers displayed above are based on latest data collected. For more information visit www.intechopen.com Chapter Amino Acids Profiling for the Diagnosis of Metabolic Disorders Yana Sandlers Abstract Inborn errors of metabolism (IEM) represent a group of inherited diseases in which genetic defect leads to the block on a metabolic pathway, resulting in a single enzyme dysfunction. -

Exome Sequencing Enhanced Package Department of Pathology and Laboratory Medicine Feb 2012 UCLA Molecular Diagnostics Laboratories Page:1
UCLA Health System Clinical Exome Sequencing Enhanced Package Department of Pathology and Laboratory Medicine Feb 2012 UCLA Molecular Diagnostics Laboratories Page:1 Gene_Symbol Total_coding_bp %_bp_>=10X Associated_Disease(OMIM) MARC1 1093 80% . MARCH1 1005 100% . MARC2 1797 92% . MARCH3 802 100% . MARCH4 1249 99% . MARCH5 861 96% . MARCH6 2907 100% . MARCH7 2161 100% . MARCH8 900 100% . MARCH9 1057 73% . MARCH10 2467 100% . MARCH11 1225 56% . SEPT1 1148 100% . SEPT2 1341 100% . SEPT3 1175 100% . SEPT4 1848 96% . SEPT5 1250 94% . SEPT6 1440 96% . SEPT7 1417 96% . SEPT8 1659 98% . SEPT9 2290 96% Hereditary Neuralgic Amyotrophy SEPT10 1605 98% . SEPT11 1334 98% . SEPT12 1113 100% . SEPT14 1335 100% . SEP15 518 100% . DEC1 229 100% . A1BG 1626 82% . A1CF 1956 100% . A2LD1 466 42% . A2M 4569 100% . A2ML1 4505 100% . UCLA Health System Clinical Exome Sequencing Enhanced Package Department of Pathology and Laboratory Medicine Feb 2012 UCLA Molecular Diagnostics Laboratories Page:2 Gene_Symbol Total_coding_bp %_bp_>=10X Associated_Disease(OMIM) A4GALT 1066 100% . A4GNT 1031 100% . AAAS 1705 100% Achalasia‐Addisonianism‐Alacrima Syndrome AACS 2091 94% . AADAC 1232 100% . AADACL2 1226 100% . AADACL3 1073 100% . AADACL4 1240 100% . AADAT 1342 97% . AAGAB 988 100% . AAK1 3095 100% . AAMP 1422 100% . AANAT 637 93% . AARS 3059 100% Charcot‐Marie‐Tooth Neuropathy Type 2 AARS 3059 100% Charcot‐Marie‐Tooth Neuropathy Type 2N AARS2 3050 100% . AARSD1 1902 98% . AASDH 3391 100% . AASDHPPT 954 100% . AASS 2873 100% Hyperlysinemia AATF 1731 99% . AATK 4181 78% . ABAT 1563 100% GABA‐Transaminase Deficiency ABCA1 6991 100% ABCA1‐Associated Familial High Density Lipoprotein Deficiency ABCA1 6991 100% Familial High Density Lipoprotein Deficiency ABCA1 6991 100% Tangier Disease ABCA10 4780 100% . -

Clinico-Radiological Phenotyping and Diagnostic Pathways in Childhood Neurometabolic Disorders—A Practical Introductory Guide
1230 Review Article on Pediatric Neuroradiology for Trainees and Fellows: An Updated Practical Guide Clinico-radiological phenotyping and diagnostic pathways in childhood neurometabolic disorders—a practical introductory guide Asthik Biswas1,2, Mukul Malhotra3, Kshitij Mankad4, Olivia Carney4, Felice D’Arco4, Karthik Muthusamy3, Sniya Valsa Sudhakar4 1Department of Diagnostic Imaging, The Hospital for Sick Children, Toronto, Ontario, Canada; 2Department of Medical Imaging, University of Toronto, Toronto, Ontario, Canada; 3Department of Neurology, Christian Medical College, Vellore, India; 4Neuroradiology Unit, Great Ormond Street Hospital NHS Foundation Trust, London, UK Contributions: (I) Conception and design: A Biswas, K Muthusamy, SV Sudhakar; (II) Administrative support: K Muthusamy, SV Sudhakar; (III) Provision of study materials or patients: A Biswas, K Muthusamy, SV Sudhakar; (IV) Collection and assembly of data: A Biswas, M Malhotra, K Muthusamy, SV Sudhakar; (V) Data analysis and interpretation: A Biswas, M Malhotra, K Muthusamy, SV Sudhakar; (VI) Manuscript writing: All authors; (VII) Final approval of manuscript: All authors. Correspondence to: Sniya Valsa Sudhakar. Neuroradiology Unit, Great Ormond Street Hospital NHS Foundation Trust, London, UK. Email: [email protected]. Abstract: Inborn errors of metabolism (IEM) although individually rare, together constitute a significant proportion of childhood neurological disorders. Majority of these disorders occur due to deficiency of an enzyme in a specific metabolic pathway, leading to damage by accumulation of a toxic substrate or deficiency of an essential metabolite. Early diagnosis is crucial in many of these conditions to prevent or minimise brain damage. Whilst many of the neuroimaging features are nonspecific, certain disorders demonstrate specific patterns due to selective vulnerability of different structures to different insults. -

Treatment of Microbial Infections with NTBC (OTT ID 1133)
Treatment of Microbial Infections with NTBC (OTT ID 1133) Inventor: Graham Moran, Ph.D., Department of Chemistry and Biochemistry, University of Wisconsin-Milwaukee Investigator: Sonia Bardy, Ph.D.; Department of Biology, University of Wisconsin-Milwaukee For further information please contact: Jessica M. Silvaggi, Ph.D. Licensing Manager UWM Research Foundation 1440 East North Avenue Milwaukee, WI 53202 Tel: 414-906-4654 [email protected] ©UWMRF 2017 1 5/18/17 Treatment of Microbial Infections with NTBC 2-(2-nitro-4-trifluoromethylbenzoyl)-1,3-cyclohexanedione Current problems with antimicrobials • Drug resistant fungi and bacteria • Lack of success with single drug treatments for infection • Fungi are difficult to treat and infections may recur • Low spectrum activity of commercial drugs • Toxicity of current commercial drugs • Unfavorable interactions of current drugs with other medications • Decreased investment in creation of novel antimicrobials Solutions and Advantages of NTBC • Excellent toxicity profile in humans • Can be used orally or topically • Bacterial infections and systemic and topical fungal infections • Great potential for use in combination with current antimicrobials • More stable in humans; longer half-life after oral ingestion • Less time to market; already FDA approved • Potential for treatment of animal infections in veterinary or livestock practices ©UWMRF 2017 2 5/18/17 Market Potential and Intellectual Property Market • According to the CDC, over 70% of hospital bacterial infections are resistant to one or more classes of antibacterial drugs • In the U.S. alone these infections result in $4.5 billion in excess healthcare costs • The antibacterial market is expected to reach $43.8 billion in 2016 (Visiongain) • Revenues for systemic antifungal drugs are predicted to approach $6 billion by 2014 (Datamonitor) • Antibiotic resistance in fungi and bacteria has become increasingly prevalent leading to a lack of success with single drug treatments Intellectual Property • A notice of allowance has been received for U.S.