A Network-Based Gene-Weighting Approach for Pathway Analysis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Association Between Chymase Gene Polymorphisms and Atrial Fibrillation
Zhou et al. BMC Cardiovascular Disorders (2019) 19:321 https://doi.org/10.1186/s12872-019-01300-7 RESEARCH ARTICLE Open Access Association between chymase gene polymorphisms and atrial fibrillation in Chinese Han population Dongchen Zhou, Yuewei Chen, Jiaxin Wu, Jiabo Shen, Yushan Shang, Liangrong Zheng and Xudong Xie* Abstract Background: Chymase is the major angiotensin II (Ang II)-forming enzyme in cardiovascular tissue, with an important role in atrial remodeling. This study aimed to examine the association between chymase 1 gene (CMA1) polymorphisms and atrial fibrillation (AF) in a Chinese Han population. Methods: This case-control study enrolled 126 patients with lone AF and 120 age- and sex-matched healthy controls, all from a Chinese Han population. Five CMA1 polymorphisms were genotyped. Results: The CMA1 polymorphism rs1800875 (G-1903A) was associated with AF. The frequency of the GG genotype was significantly higher in AF patients compared with controls (p = 0.009). Haplotype analysis further demonstrated an increased risk of AF associated with the rs1800875-G haplotype (Hap8 TGTTG, odds ratio (OR) = 1.668, 95% CI 1.132–2.458, p = 0.009), and a decreased risk for the rs1800875-A haplotype (Hap5 TATTG, OR = 0.178, 95% CI 0.042– 0.749, p = 0.008). Conclusions: CMA1 polymorphisms may be associated with AF, and the rs1800875 GG genotype might be a susceptibility factor for AF in the Chinese Han population. Keywords: Atrial fibrillation, CMA1, Chymase, Single nucleotide polymorphism, Angiotensin II Background ‘lone AF’, occur in the absence of identifiable underlying Atrial fibrillation (AF) is the most common type of sus- cardiovascular or other comorbid diseases [4]. -

CMA1 Rabbit Polyclonal Antibody – TA342809 | Origene
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for TA342809 CMA1 Rabbit Polyclonal Antibody Product data: Product Type: Primary Antibodies Applications: WB Recommended Dilution: WB Reactivity: Human Host: Rabbit Isotype: IgG Clonality: Polyclonal Immunogen: The immunogen for anti-CMA1 antibody: synthetic peptide directed towards the C terminal of human CMA1. Synthetic peptide located within the following region: EVKLRLMDPQACSHFRDFDHNLQLCVGNPRKTKSAFKGDSGGPLLCAGVA Formulation: Liquid. Purified antibody supplied in 1x PBS buffer with 0.09% (w/v) sodium azide and 2% sucrose. Note that this product is shipped as lyophilized powder to China customers. Conjugation: Unconjugated Storage: Store at -20°C as received. Stability: Stable for 12 months from date of receipt. Predicted Protein Size: 27 kDa Gene Name: chymase 1 Database Link: NP_001827 Entrez Gene 1215 Human P23946 Background: CMA1 is a chymotryptic serine proteinase that belongs to the peptidase family S1. It is expressed in mast cells and thought to function in the degradation of the extracellular matrix, the regulation of submucosal gland secretion, and the generation of vasoactive peptides. In the heart and blood vessels, this protein, rather than angiotensin converting enzyme, is largely responsible for converting angiotensin I to the vasoactive peptide angiotensin II. Angiotensin II has been implicated in blood pressure control and in the pathogenesis of hypertension, cardiac hypertrophy, and heart failure. Thus, this gene product is a target for cardiovascular disease therapies. This product is to be used for laboratory only. Not for diagnostic or therapeutic use. -

Based on Network Pharmacology Tools to Investigate the Molecular Mechanism of Cordyceps Sinensis on the Treatment of Diabetic Nephropathy
Hindawi Journal of Diabetes Research Volume 2021, Article ID 8891093, 12 pages https://doi.org/10.1155/2021/8891093 Research Article Based on Network Pharmacology Tools to Investigate the Molecular Mechanism of Cordyceps sinensis on the Treatment of Diabetic Nephropathy Yan Li,1 Lei Wang,2 Bojun Xu,1 Liangbin Zhao,1 Li Li,1 Keyang Xu,3 Anqi Tang,1 Shasha Zhou,1 Lu Song,1 Xiao Zhang,1 and Huakui Zhan 1 1Hospital of Chengdu University of Traditional Chinese Medicine, Chengdu, 610072 Sichuan, China 2Key Laboratory of Chinese Internal Medicine of Ministry of Education and Dongzhimen Hospital, Beijing University of Chinese Medicine, Beijing 100700, China 3Zhejiang Chinese Medical University, Hangzhou, 310053 Zhejiang, China Correspondence should be addressed to Huakui Zhan; [email protected] Received 27 August 2020; Revised 17 January 2021; Accepted 24 January 2021; Published 8 February 2021 Academic Editor: Michaelangela Barbieri Copyright © 2021 Yan Li et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Background. Diabetic nephropathy (DN) is one of the most common complications of diabetes mellitus and is a major cause of end- stage kidney disease. Cordyceps sinensis (Cordyceps, Dong Chong Xia Cao) is a widely applied ingredient for treating patients with DN in China, while the molecular mechanisms remain unclear. This study is aimed at revealing the therapeutic mechanisms of Cordyceps in DN by undertaking a network pharmacology analysis. Materials and Methods. In this study, active ingredients and associated target proteins of Cordyceps sinensis were obtained via Traditional Chinese Medicine Systems Pharmacology Database (TCMSP) and Swiss Target Prediction platform, then reconfirmed by using PubChem databases. -
Primepcr™Assay Validation Report
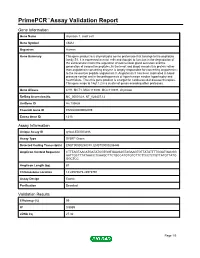
PrimePCR™Assay Validation Report Gene Information Gene Name chymase 1, mast cell Gene Symbol CMA1 Organism Human Gene Summary This gene product is a chymotryptic serine proteinase that belongs to the peptidase family S1. It is expressed in mast cells and thought to function in the degradation of the extracellular matrix the regulation of submucosal gland secretion and the generation of vasoactive peptides. In the heart and blood vessels this protein rather than angiotensin converting enzyme is largely responsible for converting angiotensin I to the vasoactive peptide angiotensin II. Angiotensin II has been implicated in blood pressure control and in the pathogenesis of hypertension cardiac hypertrophy and heart failure. Thus this gene product is a target for cardiovascular disease therapies. This gene maps to 14q11.2 in a cluster of genes encoding other proteases. Gene Aliases CYH, MCT1, MGC119890, MGC119891, chymase RefSeq Accession No. NC_000014.8, NT_026437.12 UniGene ID Hs.135626 Ensembl Gene ID ENSG00000092009 Entrez Gene ID 1215 Assay Information Unique Assay ID qHsaCED0003486 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000250378, ENST00000206446 Amplicon Context Sequence CTTTAGTAACATGATATCGTGGTGAAGAGTAGAAGTGTTATATTTTGGATGACGG AATTGCTTTATAACCTCAAGCTTCTGCCATGTGTCTTCTTCCTCTGTTATGTTATG GGCTCC Amplicon Length (bp) 87 Chromosome Location 14:24975675-24975791 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 99 R2 0.9989 cDNA Cq 27.32 Page 1/5 PrimePCR™Assay Validation Report cDNA Tm (Celsius) 78 -

Gene Expression Profiling of Lymph Node Sub-Capsular Sinus Macrophages in Cancer
ORIGINAL RESEARCH published: 08 June 2021 doi: 10.3389/fimmu.2021.672123 Gene Expression Profiling of Lymph Node Sub-Capsular Sinus Macrophages in Cancer † † Danilo Pellin 1 , Natalie Claudio 2,3 , Zihan Guo 2,4, Tahereh Ziglari 2 and Ferdinando Pucci 2,3* 1 Gene Therapy Program, Dana-Farber/Boston Children’s Cancer and Blood Disorders Center, Harvard Medical School, Boston, MA, United States, 2 Department of Otolaryngology – Head and Neck Surgery, Oregon Health and Science University, Portland, OR, United States, 3 Department of Cell, Developmental & Cancer Biology, Oregon Health and Science University, Portland, OR, United States, 4 Program in Cancer Biology, Oregon Health and Science University, Portland, OR, United States Lymph nodes are key lymphoid organs collecting lymph fluid and migratory cells from the tissue area they survey. When cancerous cells arise within a tissue, the sentinel lymph node is the first immunological organ to mount an immune response. Sub-capsular sinus Edited by: Karine Rachel Prudent Breckpot, macrophages (SSMs) are specialized macrophages residing in the lymph nodes that play Vrije University Brussel, Belgium important roles as gatekeepers against particulate antigenic material. In the context of Reviewed by: cancer, SSMs capture tumor-derived extracellular vesicles (tEVs), a form of particulate Ioannis S. Pateras, National and Kapodistrian University of antigen released in high amounts by tumor cells. We and others have recently Athens, Greece demonstrated that SSMs possess anti-tumor activity because in their absence tumors Antonio Giovanni Solimando, progress faster. A comprehensive profiling of SSMs represents an important first step to University of Bari Aldo Moro, Italy Rohit Singh, identify the cellular and molecular mechanisms responsible for SSM anti-tumor activity. -

Genetic Susceptibility to Atopic Dermatitis Chikako Kiyohara1, Keiko Tanaka2 and Yoshihiro Miyake2
Allergology International. 2008;57:39-56 ! DOI: 10.2332 allergolint.R-07-150 REVIEW ARTICLE Genetic Susceptibility to Atopic Dermatitis Chikako Kiyohara1, Keiko Tanaka2 and Yoshihiro Miyake2 ABSTRACT Atopic dermatitis (AD) is a chronic inflammatory skin disorder with an increasing prevalence in industrialized countries. AD belongs to the group of allergic disorders that includes food allergy, allergic rhinitis, and asthma. A multifactorial background for AD has been suggested, with genetic as well as environmental factors influenc- ing disease development. Recent breakthroughs in genetic methodology have greatly augmented our under- standing of the contribution of genetics to susceptibility to AD. A candidate gene association study is a general approach to identify susceptibility genes. Fifty three candidate gene studies (50 genes) have identified 19 genes associated with AD risk in at least one study. Significant associations between single nucleotide poly- morphisms (SNPs) in chemokines (chymase 1―1903A > G), cytokines (interleukin13 Arg144Gln), cytokine re- ceptors (interleukin 4 receptor 1727 G > A) and SPINK 1258 G > A have been replicated in more than one studies. These SNPs may be promising for identifying at-risk individuals. SNPs, even those not strongly associ- ated with AD, should be considered potentially important because AD is a common disease. Even a small in- crease in risk can translate to a large number of AD cases. Consortia and international collaborative studies, which may maximize study efficacy and overcome the limitations of individual studies, are needed to help fur- ther illuminate the complex landscape of AD risk and genetic variations. KEY WORDS atopic dermatitis, epidemiology, genetic polymorphism with asthma and allergic rhinitis, atopic dermatitis is INTRODUCTION an important manifestation of atopy that is character- The atopic diseases, particularly atopic dermatitis, ized by the formation of allergy antibodies (IgE) to food allergy, asthma and hay fever, are among the environmental allergens. -

Gene Expression Profiling of Human Mast Cell Subtypes
Allergology International. 2006;55:173-179 ORIGINAL ARTICLE Gene Expression Profiling of Human Mast Cell Subtypes: An In Silico Study Hirohisa Saito1,2, Kenji Matsumoto1, Shigeru Okumura2, Jun-ichi Kashiwakura2, Keisuke Oboki2, Hidenori Yokoi2, Naotomo Kambe3, Ken Ohta4 and Yoshimichi Okayama2 ABSTRACT Background: Human mast cells (MCs) were classified into at least two subtypes, i.e., tryptase- and chymase- positive MCs (MCTC) and tryptase-only-positive MCs (MCT). However, differences in global molecular expres- sion between these subtypes are unknown. Methods: We analyzed public microarray data of MC subtypes derived from various tissues and those of pe- ripheral blood granulocytes by using hierarchical clustering methods to understand the global gene expression profiles. Results: All the transcripts subjected to this clustering analysis were classified into two large clusters, i.e., MC- preferential or granulocyte-preferential. In the original works, MCs from tonsil, lung and skin had been cultured for more than several weeks to obtain highly viable and pure cell populations, and these MCs retained their typical profiles such as intensities of chymase protein expression. Most of the transcripts were commonly ex- pressed by these MC subtypes. However, tonsil-derived MCs and skin-derived MCs but not lung-derived MCs expressed high levels of chymase (CMA1) as expected for the properties of MCTC and MCT. These CMA1-high MCs and CMA1-low MCs respectively expressed distinct sets of transcripts as small gene clusters as well as CMA-1 even after being cultured in the absence of a tissue environment. Conclusions: The MC lineage seems to be far from the granulocyte lineages including basophils. -

Comprehensive Analysis Reveals Novel Gene Signature in Head and Neck Squamous Cell Carcinoma: Predicting Is Associated with Poor Prognosis in Patients
5892 Original Article Comprehensive analysis reveals novel gene signature in head and neck squamous cell carcinoma: predicting is associated with poor prognosis in patients Yixin Sun1,2#, Quan Zhang1,2#, Lanlin Yao2#, Shuai Wang3, Zhiming Zhang1,2 1Department of Breast Surgery, The First Affiliated Hospital of Xiamen University, School of Medicine, Xiamen University, Xiamen, China; 2School of Medicine, Xiamen University, Xiamen, China; 3State Key Laboratory of Cellular Stress Biology, School of Life Sciences, Xiamen University, Xiamen, China Contributions: (I) Conception and design: Y Sun, Q Zhang; (II) Administrative support: Z Zhang; (III) Provision of study materials or patients: Y Sun, Q Zhang; (IV) Collection and assembly of data: Y Sun, L Yao; (V) Data analysis and interpretation: Y Sun, S Wang; (VI) Manuscript writing: All authors; (VII) Final approval of manuscript: All authors. #These authors contributed equally to this work. Correspondence to: Zhiming Zhang. Department of Surgery, The First Affiliated Hospital of Xiamen University, Xiamen, China. Email: [email protected]. Background: Head and neck squamous cell carcinoma (HNSC) remains an important public health problem, with classic risk factors being smoking and excessive alcohol consumption and usually has a poor prognosis. Therefore, it is important to explore the underlying mechanisms of tumorigenesis and screen the genes and pathways identified from such studies and their role in pathogenesis. The purpose of this study was to identify genes or signal pathways associated with the development of HNSC. Methods: In this study, we downloaded gene expression profiles of GSE53819 from the Gene Expression Omnibus (GEO) database, including 18 HNSC tissues and 18 normal tissues. -

Development of Network-Based Analysis Methods with Application to the Genetic Component of Asthma Yuanlong Liu
Development of network-based analysis methods with application to the genetic component of asthma Yuanlong Liu To cite this version: Yuanlong Liu. Development of network-based analysis methods with application to the genetic com- ponent of asthma. Human genetics. Université Sorbonne Paris Cité, 2017. English. NNT : 2017US- PCC329. tel-02466418 HAL Id: tel-02466418 https://tel.archives-ouvertes.fr/tel-02466418 Submitted on 4 Feb 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Thèse de doctorat de l’Université Sorbonne Paris Cité Préparé à l’Université Paris Diderot ÉCOLE DOCTORALE PIERRE LOUIS DE SANTÉ PUBLIQUE À PARIS ÉPIDÉMIOLOGIE ET SCIENCES DE L’INFORMATION BIOMÉDICALE (ED 393) Unité de recherche: UMR 946 - Variabilité Génétique et Maladies Humaines DOCTORAT Spécialité: Epidémiologie Génétique Yuanlong LIU Development of network-based analysis methods with application to the genetic component of asthma Thèse dirigée par Florence DEMENAIS Présentée et soutenue publiquement à Paris le 13 Novembre 2017 JURY M. Bertram MÜLLER-MYHSOK Professeur, Technische Universität München Rapporteur Mme Kristel VAN STEEN Professeur, Université de Liège Rapporteur M. Benno SCHWIKOWSKI Directeur de Recherche, Institut Pasteur Examinateur M. Mohamed NADIF Professeur, Université Paris-Descartes Examinateur M. -

Position Effect Leading to Haploinsufficiency in a Mosaic Ring Chromosome 14 in a Boy with Autism
European Journal of Human Genetics (2008) 16, 1187–1192 & 2008 Macmillan Publishers Limited All rights reserved 1018-4813/08 $32.00 www.nature.com/ejhg ARTICLE Position effect leading to haploinsufficiency in a mosaic ring chromosome 14 in a boy with autism Dries Castermans1, Bernard Thienpont2, Karolien Volders1, An Crepel2, Joris R Vermeesch2, Connie T Schrander-Stumpel3, Wim JM Van de Ven4, Jean G Steyaert2,3,5, John WM Creemers1 and Koen Devriendt*,2 1Department for Human Genetics, Laboratory for Biochemical Neuroendocrinology, University of Leuven, Belgium; 2Division of Clinical Genetics, Center for Human Genetics, University of Leuven, Belgium; 3Department of Clinical Genetics, Academic Hospital Maastricht, Research Institute Growth and Development (GROW), Maastricht University, The Netherlands; 4Department for Human Genetics, Laboratory for Molecular Oncology, University of Leuven, Belgium; 5Department of Child Psychiatry, University of Leuven, Belgium We describe an individual with autism and a coloboma of the eye carrying a mosaicism for a ring chromosome consisting of an inverted duplication of proximal chromosome 14. Of interest, the ring formation was associated with silencing of the amisyn gene present in two copies on the ring chromosome and located at 300 kb from the breakpoint. This observation lends further support for a locus for autism on proximal chromosome 14. Moreover, this case suggests that position effects need to be taken into account, when analyzing genotype–phenotype correlations based on chromosomal imbalances. European Journal of Human Genetics (2008) 16, 1187–1192; doi:10.1038/ejhg.2008.71; published online 16 April 2008 Keywords: autism; mosaic ring chromosome 14; position effect; amisyn Introduction in a mosaic state. -

An Update on the Tissue Renin Angiotensin System and Its Role in Physiology and Pathology
Journal of Cardiovascular Development and Disease Review An Update on the Tissue Renin Angiotensin System and Its Role in Physiology and Pathology Ali Nehme 1 , Fouad A. Zouein 2 , Zeinab Deris Zayeri 3 and Kazem Zibara 4,* 1 EA4173, Functional genomics of arterial hypertension, Univeristy Claude Bernard Lyon-1 (UCBL-1), 69008 Lyon, France; [email protected] 2 Department of Pharmacology and Toxicology, Heart Repair Division, Faculty of Medicine, American University of Beirut, Beirut 11-0236, Lebanon; [email protected] 3 Thalassemia & Hemoglobinopathy Research Center, Health Research Institute, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran; [email protected] 4 PRASE, Biology Department, Faculty of Sciences-I, Lebanese University, Beirut, Lebanon * Correspondence: [email protected] Received: 10 February 2019; Accepted: 26 March 2019; Published: 29 March 2019 Abstract: In its classical view, the renin angiotensin system (RAS) was defined as an endocrine system involved in blood pressure regulation and body electrolyte balance. However, the emerging concept of tissue RAS, along with the discovery of new RAS components, increased the physiological and clinical relevance of the system. Indeed, RAS has been shown to be expressed in various tissues where alterations in its expression were shown to be involved in multiple diseases including atherosclerosis, cardiac hypertrophy, type 2 diabetes (T2D) and renal fibrosis. In this chapter, we describe the new components of RAS, their tissue-specific expression, and their alterations under pathological conditions, which will help achieve more tissue- and condition-specific treatments. Keywords: renin-angiotensin-aldosterone system; tissue; expression; physiology 1. Introduction In its classical view, RAS was defined as an endocrine system involved in blood pressure regulation and body electrolyte balance. -

The Common Marmoset Genome Provides Insight Into Primate Biology and Evolution the Marmoset Genome Sequencing and Analysis Consortium*
ARTICLES OPEN The common marmoset genome provides insight into primate biology and evolution The Marmoset Genome Sequencing and Analysis Consortium* We report the whole-genome sequence of the common marmoset (Callithrix jacchus). The 2.26-Gb genome of a female marmoset was assembled using Sanger read data (6×) and a whole-genome shotgun strategy. A first analysis has permitted comparison with the genomes of apes and Old World monkeys and the identification of specific features that might contribute to the unique biology of this diminutive primate, including genetic changes that may influence body size, frequent twinning and chimerism. We observed positive selection in growth hormone/insulin-like growth factor genes (growth pathways), respiratory complex I genes (metabolic pathways), and genes encoding immunobiological factors and proteases (reproductive and immunity pathways). In addition, both protein-coding and microRNA genes related to reproduction exhibited evidence of rapid sequence evolution. This genome sequence for a New World monkey enables increased power for comparative analyses among available primate genomes and facilitates biomedical research application. Apparently unique among mammals, marmosets routinely produce positive selection in specific genes and significant conservation dizygotic twins that exchange hematopoietic stem cells in utero, a of previously unidentified segments of noncoding DNA. The mar- process that leads to lifelong chimerism1,2. As a result of this placental moset genome displays a number of unique features, such as rapid exchange, the blood of adult marmosets normally contains a substan- changes in microRNAs (miRNAs) expressed in placenta and nonsyn- tial proportion of leukocytes that are not derived from the inherited onymous changes in protein-coding genes involved in reproductive germ line of the sampled individual but rather were acquired in utero physiology, which might be related to the frequent twinning and/or from its co-twin.