Proteomic Profiling Identifies Markers for Inflammation-Related Tumor
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Mechanisms Governing Anaphylaxis: Inflammatory Cells, Mediators
International Journal of Molecular Sciences Review Mechanisms Governing Anaphylaxis: Inflammatory Cells, Mediators, Endothelial Gap Junctions and Beyond Samantha Minh Thy Nguyen 1, Chase Preston Rupprecht 2, Aaisha Haque 3, Debendra Pattanaik 4, Joseph Yusin 5 and Guha Krishnaswamy 1,3,* 1 Department of Medicine, Wake Forest School of Medicine, Winston-Salem, NC 27106, USA; [email protected] 2 The Rowan School of Osteopathic Medicine, Stratford, NJ 08084, USA; [email protected] 3 The Bill Hefner VA Medical Center, Salisbury, NC 27106, USA; [email protected] 4 Division of Allergy and Immunology, UT Memphis College of Medicine, Memphis, TN 38103, USA; [email protected] 5 The Division of Allergy and Immunology, Greater Los Angeles VA Medical Center, Los Angeles, CA 90011, USA; [email protected] * Correspondence: [email protected] Abstract: Anaphylaxis is a severe, acute, life-threatening multisystem allergic reaction resulting from the release of a plethora of mediators from mast cells culminating in serious respiratory, cardiovascular and mucocutaneous manifestations that can be fatal. Medications, foods, latex, exercise, hormones (progesterone), and clonal mast cell disorders may be responsible. More recently, novel syndromes such as delayed reactions to red meat and hereditary alpha tryptasemia have been described. Anaphylaxis manifests as sudden onset urticaria, pruritus, flushing, erythema, Citation: Nguyen, S.M.T.; Rupprecht, angioedema (lips, tongue, airways, periphery), myocardial dysfunction (hypovolemia, distributive -

Association Between Chymase Gene Polymorphisms and Atrial Fibrillation
Zhou et al. BMC Cardiovascular Disorders (2019) 19:321 https://doi.org/10.1186/s12872-019-01300-7 RESEARCH ARTICLE Open Access Association between chymase gene polymorphisms and atrial fibrillation in Chinese Han population Dongchen Zhou, Yuewei Chen, Jiaxin Wu, Jiabo Shen, Yushan Shang, Liangrong Zheng and Xudong Xie* Abstract Background: Chymase is the major angiotensin II (Ang II)-forming enzyme in cardiovascular tissue, with an important role in atrial remodeling. This study aimed to examine the association between chymase 1 gene (CMA1) polymorphisms and atrial fibrillation (AF) in a Chinese Han population. Methods: This case-control study enrolled 126 patients with lone AF and 120 age- and sex-matched healthy controls, all from a Chinese Han population. Five CMA1 polymorphisms were genotyped. Results: The CMA1 polymorphism rs1800875 (G-1903A) was associated with AF. The frequency of the GG genotype was significantly higher in AF patients compared with controls (p = 0.009). Haplotype analysis further demonstrated an increased risk of AF associated with the rs1800875-G haplotype (Hap8 TGTTG, odds ratio (OR) = 1.668, 95% CI 1.132–2.458, p = 0.009), and a decreased risk for the rs1800875-A haplotype (Hap5 TATTG, OR = 0.178, 95% CI 0.042– 0.749, p = 0.008). Conclusions: CMA1 polymorphisms may be associated with AF, and the rs1800875 GG genotype might be a susceptibility factor for AF in the Chinese Han population. Keywords: Atrial fibrillation, CMA1, Chymase, Single nucleotide polymorphism, Angiotensin II Background ‘lone AF’, occur in the absence of identifiable underlying Atrial fibrillation (AF) is the most common type of sus- cardiovascular or other comorbid diseases [4]. -

Supplementary Table 2 Gene Sets Used in GSEA
Supplementary Table 2 Gene sets used in GSEA Up in RNAi and Sign Confirmed in Inducible Gene Probe Set ID Accession Symbol Gene Title 200660_at NM_005620 S100A11 S100 calcium binding protein A11 (calgizzarin) 200785_s_at NM_002332 LRP1 low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor) 201325_s_at NM_001423 EMP1 epithelial membrane protein 1 201373_at NM_000445 PLEC1 plectin 1, intermediate filament binding protein 500kDa 201466_s_at NM_002228 JUN v-jun sarcoma virus 17 oncogene homolog (avian) 201952_at AA156721 ALCAM activated leukocyte cell adhesion molecule 202042_at NM_002109 HARS histidyl-tRNA synthetase 202074_s_at NM_021980 OPTN optineurin 202087_s_at NM_001912 CTSL cathepsin L 202588_at NM_000476 AK1 adenylate kinase 1 202609_at NM_004447 EPS8 epidermal growth factor receptor pathway substrate 8 202733_at NM_004199 P4HA2 procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha polypeptide II 202756_s_at NM_002081 GPC1 glypican 1 202786_at NM_013233 STK39 serine threonine kinase 39 (STE20/SPS1 homolog, yeast) 202859_x_at NM_000584 IL8 interleukin 8 203083_at NM_003247 THBS2 thrombospondin 2 203186_s_at NM_002961 S100A4 S100 calcium binding protein A4 (calcium protein, calvasculin, metastasin, murine placental homolog) 203232_s_at NM_000332 ATXN1 ataxin 1 203233_at NM_000418 IL4R interleukin 4 receptor 203771_s_at AA740186 BLVRA biliverdin reductase A 203821_at NM_001945 HBEGF heparin-binding EGF-like growth factor 203939_at NM_002526 NT5E 5'-nucleotidase, ecto (CD73) 203955_at NM_014811 -

Analysis of the Indacaterol-Regulated Transcriptome in Human Airway
Supplemental material to this article can be found at: http://jpet.aspetjournals.org/content/suppl/2018/04/13/jpet.118.249292.DC1 1521-0103/366/1/220–236$35.00 https://doi.org/10.1124/jpet.118.249292 THE JOURNAL OF PHARMACOLOGY AND EXPERIMENTAL THERAPEUTICS J Pharmacol Exp Ther 366:220–236, July 2018 Copyright ª 2018 by The American Society for Pharmacology and Experimental Therapeutics Analysis of the Indacaterol-Regulated Transcriptome in Human Airway Epithelial Cells Implicates Gene Expression Changes in the s Adverse and Therapeutic Effects of b2-Adrenoceptor Agonists Dong Yan, Omar Hamed, Taruna Joshi,1 Mahmoud M. Mostafa, Kyla C. Jamieson, Radhika Joshi, Robert Newton, and Mark A. Giembycz Departments of Physiology and Pharmacology (D.Y., O.H., T.J., K.C.J., R.J., M.A.G.) and Cell Biology and Anatomy (M.M.M., R.N.), Snyder Institute for Chronic Diseases, Cumming School of Medicine, University of Calgary, Calgary, Alberta, Canada Received March 22, 2018; accepted April 11, 2018 Downloaded from ABSTRACT The contribution of gene expression changes to the adverse and activity, and positive regulation of neutrophil chemotaxis. The therapeutic effects of b2-adrenoceptor agonists in asthma was general enriched GO term extracellular space was also associ- investigated using human airway epithelial cells as a therapeu- ated with indacaterol-induced genes, and many of those, in- tically relevant target. Operational model-fitting established that cluding CRISPLD2, DMBT1, GAS1, and SOCS3, have putative jpet.aspetjournals.org the long-acting b2-adrenoceptor agonists (LABA) indacaterol, anti-inflammatory, antibacterial, and/or antiviral activity. Numer- salmeterol, formoterol, and picumeterol were full agonists on ous indacaterol-regulated genes were also induced or repressed BEAS-2B cells transfected with a cAMP-response element in BEAS-2B cells and human primary bronchial epithelial cells by reporter but differed in efficacy (indacaterol $ formoterol . -

CMA1 Rabbit Polyclonal Antibody – TA342809 | Origene
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for TA342809 CMA1 Rabbit Polyclonal Antibody Product data: Product Type: Primary Antibodies Applications: WB Recommended Dilution: WB Reactivity: Human Host: Rabbit Isotype: IgG Clonality: Polyclonal Immunogen: The immunogen for anti-CMA1 antibody: synthetic peptide directed towards the C terminal of human CMA1. Synthetic peptide located within the following region: EVKLRLMDPQACSHFRDFDHNLQLCVGNPRKTKSAFKGDSGGPLLCAGVA Formulation: Liquid. Purified antibody supplied in 1x PBS buffer with 0.09% (w/v) sodium azide and 2% sucrose. Note that this product is shipped as lyophilized powder to China customers. Conjugation: Unconjugated Storage: Store at -20°C as received. Stability: Stable for 12 months from date of receipt. Predicted Protein Size: 27 kDa Gene Name: chymase 1 Database Link: NP_001827 Entrez Gene 1215 Human P23946 Background: CMA1 is a chymotryptic serine proteinase that belongs to the peptidase family S1. It is expressed in mast cells and thought to function in the degradation of the extracellular matrix, the regulation of submucosal gland secretion, and the generation of vasoactive peptides. In the heart and blood vessels, this protein, rather than angiotensin converting enzyme, is largely responsible for converting angiotensin I to the vasoactive peptide angiotensin II. Angiotensin II has been implicated in blood pressure control and in the pathogenesis of hypertension, cardiac hypertrophy, and heart failure. Thus, this gene product is a target for cardiovascular disease therapies. This product is to be used for laboratory only. Not for diagnostic or therapeutic use. -

1 No. Affymetrix ID Gene Symbol Genedescription Gotermsbp Q Value 1. 209351 at KRT14 Keratin 14 Structural Constituent of Cyto
1 Affymetrix Gene Q No. GeneDescription GOTermsBP ID Symbol value structural constituent of cytoskeleton, intermediate 1. 209351_at KRT14 keratin 14 filament, epidermis development <0.01 biological process unknown, S100 calcium binding calcium ion binding, cellular 2. 204268_at S100A2 protein A2 component unknown <0.01 regulation of progression through cell cycle, extracellular space, cytoplasm, cell proliferation, protein kinase C inhibitor activity, protein domain specific 3. 33323_r_at SFN stratifin/14-3-3σ binding <0.01 regulation of progression through cell cycle, extracellular space, cytoplasm, cell proliferation, protein kinase C inhibitor activity, protein domain specific 4. 33322_i_at SFN stratifin/14-3-3σ binding <0.01 structural constituent of cytoskeleton, intermediate 5. 201820_at KRT5 keratin 5 filament, epidermis development <0.01 structural constituent of cytoskeleton, intermediate 6. 209125_at KRT6A keratin 6A filament, ectoderm development <0.01 regulation of progression through cell cycle, extracellular space, cytoplasm, cell proliferation, protein kinase C inhibitor activity, protein domain specific 7. 209260_at SFN stratifin/14-3-3σ binding <0.01 structural constituent of cytoskeleton, intermediate 8. 213680_at KRT6B keratin 6B filament, ectoderm development <0.01 receptor activity, cytosol, integral to plasma membrane, cell surface receptor linked signal transduction, sensory perception, tumor-associated calcium visual perception, cell 9. 202286_s_at TACSTD2 signal transducer 2 proliferation, membrane <0.01 structural constituent of cytoskeleton, cytoskeleton, intermediate filament, cell-cell adherens junction, epidermis 10. 200606_at DSP desmoplakin development <0.01 lectin, galactoside- sugar binding, extracellular binding, soluble, 7 space, nucleus, apoptosis, 11. 206400_at LGALS7 (galectin 7) heterophilic cell adhesion <0.01 2 S100 calcium binding calcium ion binding, epidermis 12. 205916_at S100A7 protein A7 (psoriasin 1) development <0.01 S100 calcium binding protein A8 (calgranulin calcium ion binding, extracellular 13. -

Based on Network Pharmacology Tools to Investigate the Molecular Mechanism of Cordyceps Sinensis on the Treatment of Diabetic Nephropathy
Hindawi Journal of Diabetes Research Volume 2021, Article ID 8891093, 12 pages https://doi.org/10.1155/2021/8891093 Research Article Based on Network Pharmacology Tools to Investigate the Molecular Mechanism of Cordyceps sinensis on the Treatment of Diabetic Nephropathy Yan Li,1 Lei Wang,2 Bojun Xu,1 Liangbin Zhao,1 Li Li,1 Keyang Xu,3 Anqi Tang,1 Shasha Zhou,1 Lu Song,1 Xiao Zhang,1 and Huakui Zhan 1 1Hospital of Chengdu University of Traditional Chinese Medicine, Chengdu, 610072 Sichuan, China 2Key Laboratory of Chinese Internal Medicine of Ministry of Education and Dongzhimen Hospital, Beijing University of Chinese Medicine, Beijing 100700, China 3Zhejiang Chinese Medical University, Hangzhou, 310053 Zhejiang, China Correspondence should be addressed to Huakui Zhan; [email protected] Received 27 August 2020; Revised 17 January 2021; Accepted 24 January 2021; Published 8 February 2021 Academic Editor: Michaelangela Barbieri Copyright © 2021 Yan Li et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Background. Diabetic nephropathy (DN) is one of the most common complications of diabetes mellitus and is a major cause of end- stage kidney disease. Cordyceps sinensis (Cordyceps, Dong Chong Xia Cao) is a widely applied ingredient for treating patients with DN in China, while the molecular mechanisms remain unclear. This study is aimed at revealing the therapeutic mechanisms of Cordyceps in DN by undertaking a network pharmacology analysis. Materials and Methods. In this study, active ingredients and associated target proteins of Cordyceps sinensis were obtained via Traditional Chinese Medicine Systems Pharmacology Database (TCMSP) and Swiss Target Prediction platform, then reconfirmed by using PubChem databases. -

Transcriptomic Analysis Delineates Potential Signature Genes And
www.nature.com/scientificreports OPEN Transcriptomic analysis delineates potential signature genes and miRNAs associated with the pathogenesis of asthma Prithvi Singh1,5, Archana Sharma2,5, Rishabh Jha1, Shweta Arora2, Rafq Ahmad3, Arshad Husain Rahmani4, Saleh A. Almatroodi4, Ravins Dohare1* & Mansoor Ali Syed2* Asthma is a multifarious disease afecting several million people around the world. It has a heterogeneous risk architecture inclusive of both genetic and environmental factors. This heterogeneity can be utilised to identify diferentially expressed biomarkers of the disease, which may ultimately aid in the development of more localized and molecularly targeted therapies. In this respect, our study complies with meta-analysis of microarray datasets containing mRNA expression profles of both asthmatic and control patients, to identify the critical Diferentially Expressed Genes (DEGs) involved in the pathogenesis of asthma. We found a total of 30 DEGs out of which 13 were involved in the pathway and functional enrichment analysis. Moreover, 5 DEGs were identifed as the hub genes by network centrality-based analysis. Most hub genes were involved in protease/ antiprotease pathways. Also, 26 miRNAs and 20 TFs having an association with these hub genes were found to be intricated in a 3-node miRNA Feed-Forward Loop. Out of these, miR-34b and miR- 449c were identifed as the key miRNAs regulating the expression of SERPINB2 gene and SMAD4 transcription factor. Thus, our study is suggestive of certain miRNAs and unexplored pathways which may pave a way to unravel critical therapeutic targets in asthma. Despite recent advances in anti-asthmatic therapeutics, asthma is still a major global health concern. -
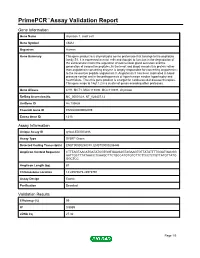
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name chymase 1, mast cell Gene Symbol CMA1 Organism Human Gene Summary This gene product is a chymotryptic serine proteinase that belongs to the peptidase family S1. It is expressed in mast cells and thought to function in the degradation of the extracellular matrix the regulation of submucosal gland secretion and the generation of vasoactive peptides. In the heart and blood vessels this protein rather than angiotensin converting enzyme is largely responsible for converting angiotensin I to the vasoactive peptide angiotensin II. Angiotensin II has been implicated in blood pressure control and in the pathogenesis of hypertension cardiac hypertrophy and heart failure. Thus this gene product is a target for cardiovascular disease therapies. This gene maps to 14q11.2 in a cluster of genes encoding other proteases. Gene Aliases CYH, MCT1, MGC119890, MGC119891, chymase RefSeq Accession No. NC_000014.8, NT_026437.12 UniGene ID Hs.135626 Ensembl Gene ID ENSG00000092009 Entrez Gene ID 1215 Assay Information Unique Assay ID qHsaCED0003486 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000250378, ENST00000206446 Amplicon Context Sequence CTTTAGTAACATGATATCGTGGTGAAGAGTAGAAGTGTTATATTTTGGATGACGG AATTGCTTTATAACCTCAAGCTTCTGCCATGTGTCTTCTTCCTCTGTTATGTTATG GGCTCC Amplicon Length (bp) 87 Chromosome Location 14:24975675-24975791 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 99 R2 0.9989 cDNA Cq 27.32 Page 1/5 PrimePCR™Assay Validation Report cDNA Tm (Celsius) 78 -

Gene Expression Profiling of Lymph Node Sub-Capsular Sinus Macrophages in Cancer
ORIGINAL RESEARCH published: 08 June 2021 doi: 10.3389/fimmu.2021.672123 Gene Expression Profiling of Lymph Node Sub-Capsular Sinus Macrophages in Cancer † † Danilo Pellin 1 , Natalie Claudio 2,3 , Zihan Guo 2,4, Tahereh Ziglari 2 and Ferdinando Pucci 2,3* 1 Gene Therapy Program, Dana-Farber/Boston Children’s Cancer and Blood Disorders Center, Harvard Medical School, Boston, MA, United States, 2 Department of Otolaryngology – Head and Neck Surgery, Oregon Health and Science University, Portland, OR, United States, 3 Department of Cell, Developmental & Cancer Biology, Oregon Health and Science University, Portland, OR, United States, 4 Program in Cancer Biology, Oregon Health and Science University, Portland, OR, United States Lymph nodes are key lymphoid organs collecting lymph fluid and migratory cells from the tissue area they survey. When cancerous cells arise within a tissue, the sentinel lymph node is the first immunological organ to mount an immune response. Sub-capsular sinus Edited by: Karine Rachel Prudent Breckpot, macrophages (SSMs) are specialized macrophages residing in the lymph nodes that play Vrije University Brussel, Belgium important roles as gatekeepers against particulate antigenic material. In the context of Reviewed by: cancer, SSMs capture tumor-derived extracellular vesicles (tEVs), a form of particulate Ioannis S. Pateras, National and Kapodistrian University of antigen released in high amounts by tumor cells. We and others have recently Athens, Greece demonstrated that SSMs possess anti-tumor activity because in their absence tumors Antonio Giovanni Solimando, progress faster. A comprehensive profiling of SSMs represents an important first step to University of Bari Aldo Moro, Italy Rohit Singh, identify the cellular and molecular mechanisms responsible for SSM anti-tumor activity. -

Clinical Impact of Inherited and Acquired Genetic Variants in Mastocytosis
International Journal of Molecular Sciences Review Clinical Impact of Inherited and Acquired Genetic Variants in Mastocytosis Boguslaw Nedoszytko 1,* , Michel Arock 2,3 , Jonathan J. Lyons 4, Guillaume Bachelot 2, Lawrence B. Schwartz 5, Andreas Reiter 6, Mohamad Jawhar 6 , Juliana Schwaab 6 , Magdalena Lange 1, Georg Greiner 7,8,9 , Gregor Hoermann 8,10 , Marek Niedoszytko 11, Dean D. Metcalfe 4 and Peter Valent 8,12 1 Department of Dermatology, Allergology and Venereology, Medical University of Gdansk, 80-211 Gdansk, Poland; [email protected] 2 Department of Hematology, APHP, Hôpital Pitié-Salpêtrière and Sorbonne University, 75013 Paris, France; [email protected] (M.A.); [email protected] (G.B.) 3 Centre de Recherche des Cordeliers, INSERM, Sorbonne Université, Université de Paris, Cell Death and Drug Resistance in Hematological Disorders Team, 75006 Paris, France 4 Laboratory of Allergic Diseases, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD 20892-188, USA; [email protected] (J.J.L.); [email protected] (D.D.M.) 5 Department of Internal Medicine, Division of Rheumatology, Allergy & Immunology, Virginia Commonwealth University, Richmond, VA 23298, USA; [email protected] 6 University Hospital Mannheim, Heidelberg University, 68167 Mannheim, Germany; [email protected] (A.R.); [email protected] (M.J.); [email protected] (J.S.) 7 Department of Laboratory Medicine, Medical -

Molecular Mechanisms Underlying Toxicant Effects on Mast Cell Signaling and Mitochondria Juyoung Katherine Shim University of Maine, [email protected]
The University of Maine DigitalCommons@UMaine Electronic Theses and Dissertations Fogler Library Summer 8-17-2018 Molecular Mechanisms Underlying Toxicant Effects on Mast Cell Signaling and Mitochondria Juyoung Katherine Shim University of Maine, [email protected] Follow this and additional works at: https://digitalcommons.library.umaine.edu/etd Part of the Molecular Biology Commons, and the Toxicology Commons Recommended Citation Shim, Juyoung Katherine, "Molecular Mechanisms Underlying Toxicant Effects on Mast Cell Signaling and Mitochondria" (2018). Electronic Theses and Dissertations. 2909. https://digitalcommons.library.umaine.edu/etd/2909 This Open-Access Thesis is brought to you for free and open access by DigitalCommons@UMaine. It has been accepted for inclusion in Electronic Theses and Dissertations by an authorized administrator of DigitalCommons@UMaine. For more information, please contact [email protected]. MOLECULAR MECHANISMS UNDERLYING TOXICANT EFFECTS ON MAST CELL SIGNALING AND MITOCHONDRIA By Juyoung K. Shim B.A. Hankook University of Foreign Studies, 1994 B.S. Bates College, 2005 A DISSERTATION Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy (in Biochemistry and Molecular Biology) The Graduate School The University of Maine August 2018 Advisory Committee: Julie A. Gosse, Associate Professor of Biochemistry; Advisor John T. Singer, Professor of Microbiology Paul J. Millard, Associate Professor of Chemical and Biological Engineering Rebecca J. Van Beneden, Professor of Biochemistry and Marine Sciences, Director for School of Marine Sciences Robert E. Gundersen, Chair of Molecular and Biomedical Sciences © 2018 Juyoung Katherine Shim All Rights Reserved ii MOLECULAR MECHANISMS UNDERLYING TOXICANT EFFECTS ON MAST CELL SIGNALING AND MITOCHONDRIA By Juyoung K.