Supplementary Table 2 Gene Sets Used in GSEA
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Seq2pathway Vignette
seq2pathway Vignette Bin Wang, Xinan Holly Yang, Arjun Kinstlick May 19, 2021 Contents 1 Abstract 1 2 Package Installation 2 3 runseq2pathway 2 4 Two main functions 3 4.1 seq2gene . .3 4.1.1 seq2gene flowchart . .3 4.1.2 runseq2gene inputs/parameters . .5 4.1.3 runseq2gene outputs . .8 4.2 gene2pathway . 10 4.2.1 gene2pathway flowchart . 11 4.2.2 gene2pathway test inputs/parameters . 11 4.2.3 gene2pathway test outputs . 12 5 Examples 13 5.1 ChIP-seq data analysis . 13 5.1.1 Map ChIP-seq enriched peaks to genes using runseq2gene .................... 13 5.1.2 Discover enriched GO terms using gene2pathway_test with gene scores . 15 5.1.3 Discover enriched GO terms using Fisher's Exact test without gene scores . 17 5.1.4 Add description for genes . 20 5.2 RNA-seq data analysis . 20 6 R environment session 23 1 Abstract Seq2pathway is a novel computational tool to analyze functional gene-sets (including signaling pathways) using variable next-generation sequencing data[1]. Integral to this tool are the \seq2gene" and \gene2pathway" components in series that infer a quantitative pathway-level profile for each sample. The seq2gene function assigns phenotype-associated significance of genomic regions to gene-level scores, where the significance could be p-values of SNPs or point mutations, protein-binding affinity, or transcriptional expression level. The seq2gene function has the feasibility to assign non-exon regions to a range of neighboring genes besides the nearest one, thus facilitating the study of functional non-coding elements[2]. Then the gene2pathway summarizes gene-level measurements to pathway-level scores, comparing the quantity of significance for gene members within a pathway with those outside a pathway. -

Mechanisms Governing Anaphylaxis: Inflammatory Cells, Mediators
International Journal of Molecular Sciences Review Mechanisms Governing Anaphylaxis: Inflammatory Cells, Mediators, Endothelial Gap Junctions and Beyond Samantha Minh Thy Nguyen 1, Chase Preston Rupprecht 2, Aaisha Haque 3, Debendra Pattanaik 4, Joseph Yusin 5 and Guha Krishnaswamy 1,3,* 1 Department of Medicine, Wake Forest School of Medicine, Winston-Salem, NC 27106, USA; [email protected] 2 The Rowan School of Osteopathic Medicine, Stratford, NJ 08084, USA; [email protected] 3 The Bill Hefner VA Medical Center, Salisbury, NC 27106, USA; [email protected] 4 Division of Allergy and Immunology, UT Memphis College of Medicine, Memphis, TN 38103, USA; [email protected] 5 The Division of Allergy and Immunology, Greater Los Angeles VA Medical Center, Los Angeles, CA 90011, USA; [email protected] * Correspondence: [email protected] Abstract: Anaphylaxis is a severe, acute, life-threatening multisystem allergic reaction resulting from the release of a plethora of mediators from mast cells culminating in serious respiratory, cardiovascular and mucocutaneous manifestations that can be fatal. Medications, foods, latex, exercise, hormones (progesterone), and clonal mast cell disorders may be responsible. More recently, novel syndromes such as delayed reactions to red meat and hereditary alpha tryptasemia have been described. Anaphylaxis manifests as sudden onset urticaria, pruritus, flushing, erythema, Citation: Nguyen, S.M.T.; Rupprecht, angioedema (lips, tongue, airways, periphery), myocardial dysfunction (hypovolemia, distributive -
Download Validation Data
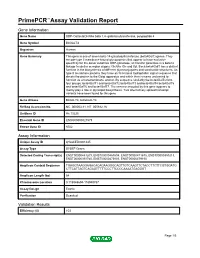
PrimePCR™Assay Validation Report Gene Information Gene Name UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 Gene Symbol B4GALT4 Organism Human Gene Summary This gene is one of seven beta-14-galactosyltransferase (beta4GalT) genes. They encode type II membrane-bound glycoproteins that appear to have exclusive specificity for the donor substrate UDP-galactose; all transfer galactose in a beta14 linkage to similar acceptor sugars: GlcNAc Glc and Xyl. Each beta4GalT has a distinct function in the biosynthesis of different glycoconjugates and saccharide structures. As type II membrane proteins they have an N-terminal hydrophobic signal sequence that directs the protein to the Golgi apparatus and which then remains uncleaved to function as a transmembrane anchor. By sequence similarity the beta4GalTs form four groups: beta4GalT1 and beta4GalT2 beta4GalT3 and beta4GalT4 beta4GalT5 and beta4GalT6 and beta4GalT7. The enzyme encoded by this gene appears to mainly play a role in glycolipid biosynthesis. Two alternatively spliced transcript variants have been found for this gene. Gene Aliases B4Gal-T4, beta4Gal-T4 RefSeq Accession No. NC_000003.11, NT_005612.16 UniGene ID Hs.13225 Ensembl Gene ID ENSG00000121578 Entrez Gene ID 8702 Assay Information Unique Assay ID qHsaCED0001445 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000483209, ENST00000467604, ENST00000471675, ENST00000359213, ENST00000393765, ENST00000475803, ENST00000479150 Amplicon Context Sequence TGAGGTAAGGAGACACAGAAGGGCAGTTGTCAAGTTCTACCTTCTTCGTGGATG CTTCATTAGTCAGAGTTTTTCCCTTCCCCAAAATGAGGGT Amplicon Length (bp) 64 Chromosome Location 3:118948694-118948787 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 103 Page 1/5 PrimePCR™Assay Validation Report R2 0.9998 cDNA Cq 21.23 cDNA Tm (Celsius) 78 gDNA Cq 23.9 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Analysis of the Indacaterol-Regulated Transcriptome in Human Airway
Supplemental material to this article can be found at: http://jpet.aspetjournals.org/content/suppl/2018/04/13/jpet.118.249292.DC1 1521-0103/366/1/220–236$35.00 https://doi.org/10.1124/jpet.118.249292 THE JOURNAL OF PHARMACOLOGY AND EXPERIMENTAL THERAPEUTICS J Pharmacol Exp Ther 366:220–236, July 2018 Copyright ª 2018 by The American Society for Pharmacology and Experimental Therapeutics Analysis of the Indacaterol-Regulated Transcriptome in Human Airway Epithelial Cells Implicates Gene Expression Changes in the s Adverse and Therapeutic Effects of b2-Adrenoceptor Agonists Dong Yan, Omar Hamed, Taruna Joshi,1 Mahmoud M. Mostafa, Kyla C. Jamieson, Radhika Joshi, Robert Newton, and Mark A. Giembycz Departments of Physiology and Pharmacology (D.Y., O.H., T.J., K.C.J., R.J., M.A.G.) and Cell Biology and Anatomy (M.M.M., R.N.), Snyder Institute for Chronic Diseases, Cumming School of Medicine, University of Calgary, Calgary, Alberta, Canada Received March 22, 2018; accepted April 11, 2018 Downloaded from ABSTRACT The contribution of gene expression changes to the adverse and activity, and positive regulation of neutrophil chemotaxis. The therapeutic effects of b2-adrenoceptor agonists in asthma was general enriched GO term extracellular space was also associ- investigated using human airway epithelial cells as a therapeu- ated with indacaterol-induced genes, and many of those, in- tically relevant target. Operational model-fitting established that cluding CRISPLD2, DMBT1, GAS1, and SOCS3, have putative jpet.aspetjournals.org the long-acting b2-adrenoceptor agonists (LABA) indacaterol, anti-inflammatory, antibacterial, and/or antiviral activity. Numer- salmeterol, formoterol, and picumeterol were full agonists on ous indacaterol-regulated genes were also induced or repressed BEAS-2B cells transfected with a cAMP-response element in BEAS-2B cells and human primary bronchial epithelial cells by reporter but differed in efficacy (indacaterol $ formoterol . -

1 No. Affymetrix ID Gene Symbol Genedescription Gotermsbp Q Value 1. 209351 at KRT14 Keratin 14 Structural Constituent of Cyto
1 Affymetrix Gene Q No. GeneDescription GOTermsBP ID Symbol value structural constituent of cytoskeleton, intermediate 1. 209351_at KRT14 keratin 14 filament, epidermis development <0.01 biological process unknown, S100 calcium binding calcium ion binding, cellular 2. 204268_at S100A2 protein A2 component unknown <0.01 regulation of progression through cell cycle, extracellular space, cytoplasm, cell proliferation, protein kinase C inhibitor activity, protein domain specific 3. 33323_r_at SFN stratifin/14-3-3σ binding <0.01 regulation of progression through cell cycle, extracellular space, cytoplasm, cell proliferation, protein kinase C inhibitor activity, protein domain specific 4. 33322_i_at SFN stratifin/14-3-3σ binding <0.01 structural constituent of cytoskeleton, intermediate 5. 201820_at KRT5 keratin 5 filament, epidermis development <0.01 structural constituent of cytoskeleton, intermediate 6. 209125_at KRT6A keratin 6A filament, ectoderm development <0.01 regulation of progression through cell cycle, extracellular space, cytoplasm, cell proliferation, protein kinase C inhibitor activity, protein domain specific 7. 209260_at SFN stratifin/14-3-3σ binding <0.01 structural constituent of cytoskeleton, intermediate 8. 213680_at KRT6B keratin 6B filament, ectoderm development <0.01 receptor activity, cytosol, integral to plasma membrane, cell surface receptor linked signal transduction, sensory perception, tumor-associated calcium visual perception, cell 9. 202286_s_at TACSTD2 signal transducer 2 proliferation, membrane <0.01 structural constituent of cytoskeleton, cytoskeleton, intermediate filament, cell-cell adherens junction, epidermis 10. 200606_at DSP desmoplakin development <0.01 lectin, galactoside- sugar binding, extracellular binding, soluble, 7 space, nucleus, apoptosis, 11. 206400_at LGALS7 (galectin 7) heterophilic cell adhesion <0.01 2 S100 calcium binding calcium ion binding, epidermis 12. 205916_at S100A7 protein A7 (psoriasin 1) development <0.01 S100 calcium binding protein A8 (calgranulin calcium ion binding, extracellular 13. -

Transcriptomic Analysis Delineates Potential Signature Genes And
www.nature.com/scientificreports OPEN Transcriptomic analysis delineates potential signature genes and miRNAs associated with the pathogenesis of asthma Prithvi Singh1,5, Archana Sharma2,5, Rishabh Jha1, Shweta Arora2, Rafq Ahmad3, Arshad Husain Rahmani4, Saleh A. Almatroodi4, Ravins Dohare1* & Mansoor Ali Syed2* Asthma is a multifarious disease afecting several million people around the world. It has a heterogeneous risk architecture inclusive of both genetic and environmental factors. This heterogeneity can be utilised to identify diferentially expressed biomarkers of the disease, which may ultimately aid in the development of more localized and molecularly targeted therapies. In this respect, our study complies with meta-analysis of microarray datasets containing mRNA expression profles of both asthmatic and control patients, to identify the critical Diferentially Expressed Genes (DEGs) involved in the pathogenesis of asthma. We found a total of 30 DEGs out of which 13 were involved in the pathway and functional enrichment analysis. Moreover, 5 DEGs were identifed as the hub genes by network centrality-based analysis. Most hub genes were involved in protease/ antiprotease pathways. Also, 26 miRNAs and 20 TFs having an association with these hub genes were found to be intricated in a 3-node miRNA Feed-Forward Loop. Out of these, miR-34b and miR- 449c were identifed as the key miRNAs regulating the expression of SERPINB2 gene and SMAD4 transcription factor. Thus, our study is suggestive of certain miRNAs and unexplored pathways which may pave a way to unravel critical therapeutic targets in asthma. Despite recent advances in anti-asthmatic therapeutics, asthma is still a major global health concern. -
![B4GALT4 Mouse Monoclonal Antibody (Biotin Conjugated) [Clone ID: OTI8B6] Product Data](https://docslib.b-cdn.net/cover/5706/b4galt4-mouse-monoclonal-antibody-biotin-conjugated-clone-id-oti8b6-product-data-1375706.webp)
B4GALT4 Mouse Monoclonal Antibody (Biotin Conjugated) [Clone ID: OTI8B6] Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for TA807809AM B4GALT4 Mouse Monoclonal Antibody (Biotin conjugated) [Clone ID: OTI8B6] Product data: Product Type: Primary Antibodies Clone Name: OTI8B6 Applications: IHC, WB Recommended Dilution: WB 1:500, IHC 1:150 Reactivity: Human Host: Mouse Isotype: IgG1 Clonality: Monoclonal Immunogen: Human recombinant protein fragment corresponding to amino acids 40-322 of human B4GALT4(NP_003769) produced in E.coli. Formulation: PBS (PH 7.3) containing 1% BSA, 50% glycerol and 0.02% sodium azide. Concentration: 0.5 mg/ml Purification: Purified from mouse ascites fluids or tissue culture supernatant by affinity chromatography (protein A/G) Conjugation: Biotin Storage: Store at -20°C as received. Stability: Stable for 12 months from date of receipt. Predicted Protein Size: 39.9 kDa Gene Name: beta-1,4-galactosyltransferase 4 Database Link: NP_003769 Entrez Gene 8702 Human O60513 This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 3 B4GALT4 Mouse Monoclonal Antibody (Biotin conjugated) [Clone ID: OTI8B6] – TA807809AM Background: This gene is one of seven beta-1,4-galactosyltransferase (beta4GalT) genes. They encode type II membrane-bound glycoproteins that appear to have exclusive specificity for the donor substrate UDP-galactose; all transfer galactose in a beta1,4 linkage to similar acceptor sugars: GlcNAc, Glc, and Xyl. Each beta4GalT has a distinct function in the biosynthesis of different glycoconjugates and saccharide structures. -

Duke University Dissertation Template
Gene-Environment Interactions in Cardiovascular Disease by Cavin Keith Ward-Caviness Graduate Program in Computational Biology and Bioinformatics Duke University Date:_______________________ Approved: ___________________________ Elizabeth R. Hauser, Supervisor ___________________________ William E. Kraus ___________________________ Sayan Mukherjee ___________________________ H. Frederik Nijhout Dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Graduate Program in Computational Biology and Bioinformatics in the Graduate School of Duke University 2014 i v ABSTRACT Gene-Environment Interactions in Cardiovascular Disease by Cavin Keith Ward-Caviness Graduate Program in Computational Biology and Bioinformatics Duke University Date:_______________________ Approved: ___________________________ Elizabeth R. Hauser, Supervisor ___________________________ William E. Kraus ___________________________ Sayan Mukherjee ___________________________ H. Frederik Nijhout An abstract of a dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Graduate Program in Computational Biology and Bioinformatics in the Graduate School of Duke University 2014 Copyright by Cavin Keith Ward-Caviness 2014 Abstract In this manuscript I seek to demonstrate the importance of gene-environment interactions in cardiovascular disease. This manuscript contains five studies each of which contributes to our understanding of the joint impact of genetic variation -

Clinical Impact of Inherited and Acquired Genetic Variants in Mastocytosis
International Journal of Molecular Sciences Review Clinical Impact of Inherited and Acquired Genetic Variants in Mastocytosis Boguslaw Nedoszytko 1,* , Michel Arock 2,3 , Jonathan J. Lyons 4, Guillaume Bachelot 2, Lawrence B. Schwartz 5, Andreas Reiter 6, Mohamad Jawhar 6 , Juliana Schwaab 6 , Magdalena Lange 1, Georg Greiner 7,8,9 , Gregor Hoermann 8,10 , Marek Niedoszytko 11, Dean D. Metcalfe 4 and Peter Valent 8,12 1 Department of Dermatology, Allergology and Venereology, Medical University of Gdansk, 80-211 Gdansk, Poland; [email protected] 2 Department of Hematology, APHP, Hôpital Pitié-Salpêtrière and Sorbonne University, 75013 Paris, France; [email protected] (M.A.); [email protected] (G.B.) 3 Centre de Recherche des Cordeliers, INSERM, Sorbonne Université, Université de Paris, Cell Death and Drug Resistance in Hematological Disorders Team, 75006 Paris, France 4 Laboratory of Allergic Diseases, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD 20892-188, USA; [email protected] (J.J.L.); [email protected] (D.D.M.) 5 Department of Internal Medicine, Division of Rheumatology, Allergy & Immunology, Virginia Commonwealth University, Richmond, VA 23298, USA; [email protected] 6 University Hospital Mannheim, Heidelberg University, 68167 Mannheim, Germany; [email protected] (A.R.); [email protected] (M.J.); [email protected] (J.S.) 7 Department of Laboratory Medicine, Medical -

Molecular Mechanisms Underlying Toxicant Effects on Mast Cell Signaling and Mitochondria Juyoung Katherine Shim University of Maine, [email protected]
The University of Maine DigitalCommons@UMaine Electronic Theses and Dissertations Fogler Library Summer 8-17-2018 Molecular Mechanisms Underlying Toxicant Effects on Mast Cell Signaling and Mitochondria Juyoung Katherine Shim University of Maine, [email protected] Follow this and additional works at: https://digitalcommons.library.umaine.edu/etd Part of the Molecular Biology Commons, and the Toxicology Commons Recommended Citation Shim, Juyoung Katherine, "Molecular Mechanisms Underlying Toxicant Effects on Mast Cell Signaling and Mitochondria" (2018). Electronic Theses and Dissertations. 2909. https://digitalcommons.library.umaine.edu/etd/2909 This Open-Access Thesis is brought to you for free and open access by DigitalCommons@UMaine. It has been accepted for inclusion in Electronic Theses and Dissertations by an authorized administrator of DigitalCommons@UMaine. For more information, please contact [email protected]. MOLECULAR MECHANISMS UNDERLYING TOXICANT EFFECTS ON MAST CELL SIGNALING AND MITOCHONDRIA By Juyoung K. Shim B.A. Hankook University of Foreign Studies, 1994 B.S. Bates College, 2005 A DISSERTATION Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy (in Biochemistry and Molecular Biology) The Graduate School The University of Maine August 2018 Advisory Committee: Julie A. Gosse, Associate Professor of Biochemistry; Advisor John T. Singer, Professor of Microbiology Paul J. Millard, Associate Professor of Chemical and Biological Engineering Rebecca J. Van Beneden, Professor of Biochemistry and Marine Sciences, Director for School of Marine Sciences Robert E. Gundersen, Chair of Molecular and Biomedical Sciences © 2018 Juyoung Katherine Shim All Rights Reserved ii MOLECULAR MECHANISMS UNDERLYING TOXICANT EFFECTS ON MAST CELL SIGNALING AND MITOCHONDRIA By Juyoung K. -

The Emerging Role of Mast Cell Proteases in Asthma
REVIEW ASTHMA The emerging role of mast cell proteases in asthma Gunnar Pejler1,2 Affiliations: 1Dept of Medical Biochemistry and Microbiology, Uppsala University, Uppsala, Sweden. 2Dept of Anatomy, Physiology and Biochemistry, Swedish University of Agricultural Sciences, Uppsala, Sweden. Correspondence: Gunnar Pejler, Dept of Medical Biochemistry and Microbiology, BMC, Uppsala University, Box 582, 75123 Uppsala, Sweden. E-mail: [email protected] @ERSpublications Mast cells express large amounts of proteases, including tryptase, chymase and carboxypeptidase A3. An extensive review of how these proteases impact on asthma shows that they can have both protective and detrimental functions. http://bit.ly/2Gu1Qp2 Cite this article as: Pejler G. The emerging role of mast cell proteases in asthma. Eur Respir J 2019; 54: 1900685 [https://doi.org/10.1183/13993003.00685-2019]. ABSTRACT It is now well established that mast cells (MCs) play a crucial role in asthma. This is supported by multiple lines of evidence, including both clinical studies and studies on MC-deficient mice. However, there is still only limited knowledge of the exact effector mechanism(s) by which MCs influence asthma pathology. MCs contain large amounts of secretory granules, which are filled with a variety of bioactive compounds including histamine, cytokines, lysosomal hydrolases, serglycin proteoglycans and a number of MC-restricted proteases. When MCs are activated, e.g. in response to IgE receptor cross- linking, the contents of their granules are released to the exterior and can cause a massive inflammatory reaction. The MC-restricted proteases include tryptases, chymases and carboxypeptidase A3, and these are expressed and stored at remarkably high levels.