Gene Expression Profiling of Human Mast Cell Subtypes
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Ige-Mediated Mast Cell Activation Promotes Inflammation And
RESEARCH COMMUNICATION IgE-mediated mast cell activation promotes inflammation and cartilage destruction in osteoarthritis Qian Wang1,2†, Christin M Lepus1,2†, Harini Raghu1,2†, Laurent L Reber3‡, Mindy M Tsai3, Heidi H Wong1,2, Ericka von Kaeppler1,2, Nithya Lingampalli1,2, Michelle S Bloom1,2, Nick Hu1,2, Eileen E Elliott1,2, Francesca Oliviero4, Leonardo Punzi4, Nicholas J Giori1,5, Stuart B Goodman5, Constance R Chu1,5, Jeremy Sokolove1,2, Yoshihiro Fukuoka6, Lawrence B Schwartz6, Stephen J Galli3,7, William H Robinson1,2* 1GRECC, VA Palo Alto Health Care System, Palo Alto, United States; 2Division of Immunology and Rheumatology, Stanford University School of Medicine, Stanford, United States; 3Department of Pathology, Stanford University School of Medicine, Stanford, United States; 4Rheumatology Unit, Department of Medicine, University of Padova, Padova, Italy; 5Department of Orthopedic Surgery, Stanford University School of Medicine, Stanford, United States; 6Department of Internal Medicine, Virginia Commonwealth University School of Medicine, Richmond, United States; 7Department of Microbiology and Immunology, Stanford University School of Medicine, Stanford, United States *For correspondence: [email protected] Abstract Osteoarthritis is characterized by articular cartilage breakdown, and emerging †These authors contributed evidence suggests that dysregulated innate immunity is likely involved. Here, we performed equally to this work proteomic, transcriptomic, and electron microscopic analyses to demonstrate that mast cells are Present address: ‡Center for aberrantly activated in human and murine osteoarthritic joint tissues. Using genetic models of mast Physiopathology of Toulouse- cell deficiency, we demonstrate that lack of mast cells attenuates osteoarthritis in mice. Using Purpan (CPTP), UMR 1043, genetic and pharmacologic approaches, we show that the IgE/FceRI/Syk signaling axis is critical for University of Toulouse, INSERM, the development of osteoarthritis. -

Biological Function of Mast Cell Chymase
Biological Function of Mast Cell Chymase In vitro and in vivo studies: a thorny pathway Elena Chugunova Department of Molecular Biosciences Uppsala Doctoral thesis Swedish University of Agricultural Sciences Uppsala 2004 Acta Universitatis Agriculturae Sueciae Veterinaria 181 ISSN 1401-6257 ISBN 91-576-6680-6 © 2004 Elena Chugunova, Uppsala Tryck: SLU Service/Repro, Uppsala 2004 Abstract Chugunova, E., 2004. Biological function of mast cell chymase mMCP-4. In vitro and in vivo studies: a thorny pathway. Doctor's dissertation. ISSN 1401-6257, ISBN 91-576-6680-6 Mast cells (MCs) are key effector cells in various types of inflammatory conditions. The MC secretory granules contain inflammatory mediators such as histamine, heparin proteoglycan (PG), cytokines and various heparin-binding proteases, including tryptases, chymases and carboxypeptidase A. Previously, a mouse strain with a defect in its heparin biosynthesis was produced by targeting the gene for NDST-2 (N-deacetylase/N-sulfotransferase-2). These mice showed reduced levels of MC inflammatory mediators such as histamine and various heparin- binding proteases, including chymases, tryptases, and carboxypeptidase A. By using this mouse strain, we found that chymase in complex with heparin PG degraded fibronectin, suggesting a role for chymase in the regulation of connective tissue composition. Further, we found that chymase/heparin PG complexes degraded and thereby inactivated both thrombin and plasmin, suggesting an additional role for chymase in regulation of extravascular coagulation and fibrinolysis. However, although our findings implicated chymase in these processes, it was not possible to exclude the contribution to the observed activities by other MC components that are influenced by the knockout of NDST-2. -

Association Between Chymase Gene Polymorphisms and Atrial Fibrillation
Zhou et al. BMC Cardiovascular Disorders (2019) 19:321 https://doi.org/10.1186/s12872-019-01300-7 RESEARCH ARTICLE Open Access Association between chymase gene polymorphisms and atrial fibrillation in Chinese Han population Dongchen Zhou, Yuewei Chen, Jiaxin Wu, Jiabo Shen, Yushan Shang, Liangrong Zheng and Xudong Xie* Abstract Background: Chymase is the major angiotensin II (Ang II)-forming enzyme in cardiovascular tissue, with an important role in atrial remodeling. This study aimed to examine the association between chymase 1 gene (CMA1) polymorphisms and atrial fibrillation (AF) in a Chinese Han population. Methods: This case-control study enrolled 126 patients with lone AF and 120 age- and sex-matched healthy controls, all from a Chinese Han population. Five CMA1 polymorphisms were genotyped. Results: The CMA1 polymorphism rs1800875 (G-1903A) was associated with AF. The frequency of the GG genotype was significantly higher in AF patients compared with controls (p = 0.009). Haplotype analysis further demonstrated an increased risk of AF associated with the rs1800875-G haplotype (Hap8 TGTTG, odds ratio (OR) = 1.668, 95% CI 1.132–2.458, p = 0.009), and a decreased risk for the rs1800875-A haplotype (Hap5 TATTG, OR = 0.178, 95% CI 0.042– 0.749, p = 0.008). Conclusions: CMA1 polymorphisms may be associated with AF, and the rs1800875 GG genotype might be a susceptibility factor for AF in the Chinese Han population. Keywords: Atrial fibrillation, CMA1, Chymase, Single nucleotide polymorphism, Angiotensin II Background ‘lone AF’, occur in the absence of identifiable underlying Atrial fibrillation (AF) is the most common type of sus- cardiovascular or other comorbid diseases [4]. -

CMA1 Rabbit Polyclonal Antibody – TA342809 | Origene
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for TA342809 CMA1 Rabbit Polyclonal Antibody Product data: Product Type: Primary Antibodies Applications: WB Recommended Dilution: WB Reactivity: Human Host: Rabbit Isotype: IgG Clonality: Polyclonal Immunogen: The immunogen for anti-CMA1 antibody: synthetic peptide directed towards the C terminal of human CMA1. Synthetic peptide located within the following region: EVKLRLMDPQACSHFRDFDHNLQLCVGNPRKTKSAFKGDSGGPLLCAGVA Formulation: Liquid. Purified antibody supplied in 1x PBS buffer with 0.09% (w/v) sodium azide and 2% sucrose. Note that this product is shipped as lyophilized powder to China customers. Conjugation: Unconjugated Storage: Store at -20°C as received. Stability: Stable for 12 months from date of receipt. Predicted Protein Size: 27 kDa Gene Name: chymase 1 Database Link: NP_001827 Entrez Gene 1215 Human P23946 Background: CMA1 is a chymotryptic serine proteinase that belongs to the peptidase family S1. It is expressed in mast cells and thought to function in the degradation of the extracellular matrix, the regulation of submucosal gland secretion, and the generation of vasoactive peptides. In the heart and blood vessels, this protein, rather than angiotensin converting enzyme, is largely responsible for converting angiotensin I to the vasoactive peptide angiotensin II. Angiotensin II has been implicated in blood pressure control and in the pathogenesis of hypertension, cardiac hypertrophy, and heart failure. Thus, this gene product is a target for cardiovascular disease therapies. This product is to be used for laboratory only. Not for diagnostic or therapeutic use. -

Extended Cleavage Specificities of Two Mast Cell Chymase-Related
International Journal of Molecular Sciences Article Extended Cleavage Specificities of Two Mast Cell Chymase-Related Proteases and One Granzyme B-Like Protease from the Platypus, a Monotreme Zhirong Fu, Srinivas Akula , Michael Thorpe and Lars Hellman * Department of Cell and Molecular Biology, Uppsala University, Uppsala, The Biomedical Center, Box 596, SE-751 24 Uppsala, Sweden; [email protected] (Z.F.); [email protected] (S.A.); [email protected] (M.T.) * Correspondence: [email protected]; Tel.: +46-(0)18-471-4532; Fax: +46-(0)18-471-4862 Received: 20 November 2019; Accepted: 31 December 2019; Published: 2 January 2020 Abstract: Mast cells (MCs) are inflammatory cells primarily found in tissues in close contact with the external environment, such as the skin and the intestinal mucosa. They store large amounts of active components in cytoplasmic granules, ready for rapid release. The major protein content of these granules is proteases, which can account for up to 35 % of the total cellular protein. Depending on their primary cleavage specificity, they can generally be subdivided into chymases and tryptases. Here we present the extended cleavage specificities of two such proteases from the platypus. Both of them show an extended chymotrypsin-like specificity almost identical to other mammalian MC chymases. This suggests that MC chymotryptic enzymes have been conserved, both in structure and extended cleavage specificity, for more than 200 million years, indicating major functions in MC-dependent physiological processes. We have also studied a third closely related protease, originating from the same chymase locus whose cleavage specificity is closely related to the apoptosis-inducing protease from cytotoxic T cells, granzyme B. -

Based on Network Pharmacology Tools to Investigate the Molecular Mechanism of Cordyceps Sinensis on the Treatment of Diabetic Nephropathy
Hindawi Journal of Diabetes Research Volume 2021, Article ID 8891093, 12 pages https://doi.org/10.1155/2021/8891093 Research Article Based on Network Pharmacology Tools to Investigate the Molecular Mechanism of Cordyceps sinensis on the Treatment of Diabetic Nephropathy Yan Li,1 Lei Wang,2 Bojun Xu,1 Liangbin Zhao,1 Li Li,1 Keyang Xu,3 Anqi Tang,1 Shasha Zhou,1 Lu Song,1 Xiao Zhang,1 and Huakui Zhan 1 1Hospital of Chengdu University of Traditional Chinese Medicine, Chengdu, 610072 Sichuan, China 2Key Laboratory of Chinese Internal Medicine of Ministry of Education and Dongzhimen Hospital, Beijing University of Chinese Medicine, Beijing 100700, China 3Zhejiang Chinese Medical University, Hangzhou, 310053 Zhejiang, China Correspondence should be addressed to Huakui Zhan; [email protected] Received 27 August 2020; Revised 17 January 2021; Accepted 24 January 2021; Published 8 February 2021 Academic Editor: Michaelangela Barbieri Copyright © 2021 Yan Li et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Background. Diabetic nephropathy (DN) is one of the most common complications of diabetes mellitus and is a major cause of end- stage kidney disease. Cordyceps sinensis (Cordyceps, Dong Chong Xia Cao) is a widely applied ingredient for treating patients with DN in China, while the molecular mechanisms remain unclear. This study is aimed at revealing the therapeutic mechanisms of Cordyceps in DN by undertaking a network pharmacology analysis. Materials and Methods. In this study, active ingredients and associated target proteins of Cordyceps sinensis were obtained via Traditional Chinese Medicine Systems Pharmacology Database (TCMSP) and Swiss Target Prediction platform, then reconfirmed by using PubChem databases. -
Primepcr™Assay Validation Report
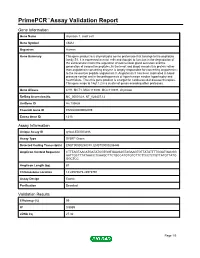
PrimePCR™Assay Validation Report Gene Information Gene Name chymase 1, mast cell Gene Symbol CMA1 Organism Human Gene Summary This gene product is a chymotryptic serine proteinase that belongs to the peptidase family S1. It is expressed in mast cells and thought to function in the degradation of the extracellular matrix the regulation of submucosal gland secretion and the generation of vasoactive peptides. In the heart and blood vessels this protein rather than angiotensin converting enzyme is largely responsible for converting angiotensin I to the vasoactive peptide angiotensin II. Angiotensin II has been implicated in blood pressure control and in the pathogenesis of hypertension cardiac hypertrophy and heart failure. Thus this gene product is a target for cardiovascular disease therapies. This gene maps to 14q11.2 in a cluster of genes encoding other proteases. Gene Aliases CYH, MCT1, MGC119890, MGC119891, chymase RefSeq Accession No. NC_000014.8, NT_026437.12 UniGene ID Hs.135626 Ensembl Gene ID ENSG00000092009 Entrez Gene ID 1215 Assay Information Unique Assay ID qHsaCED0003486 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000250378, ENST00000206446 Amplicon Context Sequence CTTTAGTAACATGATATCGTGGTGAAGAGTAGAAGTGTTATATTTTGGATGACGG AATTGCTTTATAACCTCAAGCTTCTGCCATGTGTCTTCTTCCTCTGTTATGTTATG GGCTCC Amplicon Length (bp) 87 Chromosome Location 14:24975675-24975791 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 99 R2 0.9989 cDNA Cq 27.32 Page 1/5 PrimePCR™Assay Validation Report cDNA Tm (Celsius) 78 -

Like Transmembrane Γ Evolved From
Mast Cell α and β Tryptases Changed Rapidly during Primate Speciation and Evolved from γ-Like Transmembrane Peptidases in Ancestral Vertebrates This information is current as of September 25, 2021. Neil N. Trivedi, Qiao Tong, Kavita Raman, Vikash J. Bhagwandin and George H. Caughey J Immunol 2007; 179:6072-6079; ; doi: 10.4049/jimmunol.179.9.6072 http://www.jimmunol.org/content/179/9/6072 Downloaded from References This article cites 34 articles, 15 of which you can access for free at: http://www.jimmunol.org/content/179/9/6072.full#ref-list-1 http://www.jimmunol.org/ Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication by guest on September 25, 2021 *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2007 by The American Association of Immunologists All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology Mast Cell ␣ and  Tryptases Changed Rapidly during Primate Speciation and Evolved from ␥-Like Transmembrane Peptidases in Ancestral Vertebrates1 Neil N. Trivedi, Qiao Tong, Kavita Raman, Vikash J. Bhagwandin, and George H. -

Mast Cell Chymase and Kidney Disease
International Journal of Molecular Sciences Review Mast Cell Chymase and Kidney Disease Shamila Vibhushan 1,2, Manuela Bratti 1,2 , Juan Eduardo Montero-Hernández 1,2 , Alaa El Ghoneimi 1,2,3, Marc Benhamou 1,2, Nicolas Charles 1,2 , Eric Daugas 1,2,4 and Ulrich Blank 1,2,* 1 Centre de Recherche sur l’inflammation, CNRS ERL8252, Faculté de Médecine site Bichat, Université de Paris, Inserm UMR1149, 16 rue Henri Huchard, F-75018 Paris, France; [email protected] (S.V.); [email protected] (M.B.); [email protected] (J.E.M.-H.); [email protected] (A.E.G.); [email protected] (M.B.); [email protected] (N.C.); [email protected] (E.D.) 2 Laboratoire d’Excellence Inflamex, Université de Paris, F-75018 Paris, France 3 Department of Pediatric Surgery and Urology, Hôpital Universitaire Robert Debré, Assistance Publique—Hôpitaux de Paris (APHP), F-75019 Paris, France 4 Service de Néphrologie, Groupe Hospitalier Universitaire Bichat-Claude Bernard, Assistance Publique—Hôpitaux de Paris (APHP), F-75019 Paris, France * Correspondence: [email protected] Abstract: A sizable part (~2%) of the human genome encodes for proteases. They are involved in many physiological processes, such as development, reproduction and inflammation, but also play a role in pathology. Mast cells (MC) contain a variety of MC specific proteases, the expression of which may differ between various MC subtypes. Amongst these proteases, chymase represents up to 25% of the total proteins in the MC and is released from cytoplasmic granules upon activation. -

Gene Expression Profiling of Lymph Node Sub-Capsular Sinus Macrophages in Cancer
ORIGINAL RESEARCH published: 08 June 2021 doi: 10.3389/fimmu.2021.672123 Gene Expression Profiling of Lymph Node Sub-Capsular Sinus Macrophages in Cancer † † Danilo Pellin 1 , Natalie Claudio 2,3 , Zihan Guo 2,4, Tahereh Ziglari 2 and Ferdinando Pucci 2,3* 1 Gene Therapy Program, Dana-Farber/Boston Children’s Cancer and Blood Disorders Center, Harvard Medical School, Boston, MA, United States, 2 Department of Otolaryngology – Head and Neck Surgery, Oregon Health and Science University, Portland, OR, United States, 3 Department of Cell, Developmental & Cancer Biology, Oregon Health and Science University, Portland, OR, United States, 4 Program in Cancer Biology, Oregon Health and Science University, Portland, OR, United States Lymph nodes are key lymphoid organs collecting lymph fluid and migratory cells from the tissue area they survey. When cancerous cells arise within a tissue, the sentinel lymph node is the first immunological organ to mount an immune response. Sub-capsular sinus Edited by: Karine Rachel Prudent Breckpot, macrophages (SSMs) are specialized macrophages residing in the lymph nodes that play Vrije University Brussel, Belgium important roles as gatekeepers against particulate antigenic material. In the context of Reviewed by: cancer, SSMs capture tumor-derived extracellular vesicles (tEVs), a form of particulate Ioannis S. Pateras, National and Kapodistrian University of antigen released in high amounts by tumor cells. We and others have recently Athens, Greece demonstrated that SSMs possess anti-tumor activity because in their absence tumors Antonio Giovanni Solimando, progress faster. A comprehensive profiling of SSMs represents an important first step to University of Bari Aldo Moro, Italy Rohit Singh, identify the cellular and molecular mechanisms responsible for SSM anti-tumor activity. -

Correlation of Serpin–Protease Expression by Comparative Analysis of Real-Time PCR Profiling Data
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Genomics 88 (2006) 173–184 www.elsevier.com/locate/ygeno Correlation of serpin–protease expression by comparative analysis of real-time PCR profiling data Sunita Badola a, Heidi Spurling a, Keith Robison a, Eric R. Fedyk a, Gary A. Silverman b, ⁎ Jochen Strayle c, Rosana Kapeller a,1, Christopher A. Tsu a, a Millennium Pharmaceuticals, Inc., 40 Landsdowne Street, Cambridge, MA 02139, USA b Department of Pediatrics, University of Pittsburgh School of Medicine, Magee-Women’s Hospital, 300 Halket Street, Pittsburgh, PA 15213, USA c Bayer HealthCare AG, 42096 Wuppertal, Germany Received 2 December 2005; accepted 27 March 2006 Available online 18 May 2006 Abstract Imbalanced protease activity has long been recognized in the progression of disease states such as cancer and inflammation. Serpins, the largest family of endogenous protease inhibitors, target a wide variety of serine and cysteine proteases and play a role in a number of physiological and pathological states. The expression profiles of 20 serpins and 105 serine and cysteine proteases were determined across a panel of normal and diseased human tissues. In general, expression of serpins was highly restricted in both normal and diseased tissues, suggesting defined physiological roles for these protease inhibitors. A high correlation in expression for a particular serpin–protease pair in healthy tissues was often predictive of a biological interaction. The most striking finding was the dramatic change observed in the regulation of expression between proteases and their cognate inhibitors in diseased tissues. -

TPSAB1 Monoclonal Antibody (AA1)
Website: thermofisher.com Lot Number: SB2348992 Customer Service(US): 1 800 955 6288 ext. 1 Technical Support (US): 1 800 955 6288 ext. 441 thermofisher.com/contactus TPSAB1 Monoclonal Antibody (AA1) Catalog Number:MA5-11711 Product Data Sheet Details Species Reactivity Size 500 µL Canine, Feline, Human, Tested species reactivity Non-human primate Host / Isotype Mouse / IgG1 Published Species Reactivity Human, Mouse, Rat Class Monoclonal Type Antibody Tested Applications Dilution * Clone AA1 Western Blot (WB) 1:50 Human mast cell tryptase Immunohistochemistry (Paraffin) Immunogen 1:2000 purified from human lung tissues (IHC (P)) Form Liquid Published Applications Concentration 50µg/ml Immunocytochemistry (ICC) See 1 publications below Purification Protein G Immunohistochemistry (IHC) See 16 publications below Storage Buffer PBS, pH 7.4, with 0.2% BSA * Suggested working dilutions are given as a guide only. It is recommended that the user titrate the product for use in their own experiment using appropriate negative and positive controls. Contains 0.09% sodium azide Storage Conditions 4° C Product Specific Information MA5-11711 targets Mast Cell Tryptase in IHC (P) and WB applications and shows reactivity with Canine, Feline, Human, and Non-human primate samples. The MA5-11711 immunogen is human mast cell tryptase purified from human lung tissues. Background/Target Information Mast cells contain a number of preformed chemical mediators such as histamine, chymase, carboxypeptidase and proteolytic tryptase. A substantial quantity of tryptase is estimated to be found in mast cells of skin and lung and suggestes this enzyme plays a major role in mast cell mediated events. In vitro studies indicate tryptase can cleave C3 to form C3a anaphylatoxin, inactivate fibrinogen as a coaguable substrate for thrombin and activate latent collagenase.