Comparative Analysis of Orthologous Mast Cell Chymases Active Sites Using Bioinformatics Tools
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

(12) United States Patent (10) Patent No.: US 6,395,889 B1 Robison (45) Date of Patent: May 28, 2002
USOO6395889B1 (12) United States Patent (10) Patent No.: US 6,395,889 B1 Robison (45) Date of Patent: May 28, 2002 (54) NUCLEIC ACID MOLECULES ENCODING WO WO-98/56804 A1 * 12/1998 ........... CO7H/21/02 HUMAN PROTEASE HOMOLOGS WO WO-99/0785.0 A1 * 2/1999 ... C12N/15/12 WO WO-99/37660 A1 * 7/1999 ........... CO7H/21/04 (75) Inventor: fish E. Robison, Wilmington, MA OTHER PUBLICATIONS Vazquez, F., et al., 1999, “METH-1, a human ortholog of (73) Assignee: Millennium Pharmaceuticals, Inc., ADAMTS-1, and METH-2 are members of a new family of Cambridge, MA (US) proteins with angio-inhibitory activity', The Journal of c: - 0 Biological Chemistry, vol. 274, No. 33, pp. 23349–23357.* (*) Notice: Subject to any disclaimer, the term of this Descriptors of Protease Classes in Prosite and Pfam Data patent is extended or adjusted under 35 bases. U.S.C. 154(b) by 0 days. * cited by examiner (21) Appl. No.: 09/392, 184 Primary Examiner Ponnathapu Achutamurthy (22) Filed: Sep. 9, 1999 ASSistant Examiner William W. Moore (51) Int. Cl." C12N 15/57; C12N 15/12; (74) Attorney, Agent, or Firm-Alston & Bird LLP C12N 9/64; C12N 15/79 (57) ABSTRACT (52) U.S. Cl. .................... 536/23.2; 536/23.5; 435/69.1; 435/252.3; 435/320.1 The invention relates to polynucleotides encoding newly (58) Field of Search ............................... 536,232,235. identified protease homologs. The invention also relates to 435/6, 226, 69.1, 252.3 the proteases. The invention further relates to methods using s s s/ - - -us the protease polypeptides and polynucleotides as a target for (56) References Cited diagnosis and treatment in protease-mediated disorders. -

Structure of Human Aspartyl Aminopeptidase Complexed With
Chaikuad et al. BMC Structural Biology 2012, 12:14 http://www.biomedcentral.com/1472-6807/12/14 RESEARCH ARTICLE Open Access Structure of human aspartyl aminopeptidase complexed with substrate analogue: insight into catalytic mechanism, substrate specificity and M18 peptidase family Apirat Chaikuad1, Ewa S Pilka1, Antonio De Riso2, Frank von Delft1, Kathryn L Kavanagh1, Catherine Vénien-Bryan2, Udo Oppermann1,3 and Wyatt W Yue1* Abstract Backround: Aspartyl aminopeptidase (DNPEP), with specificity towards an acidic amino acid at the N-terminus, is the only mammalian member among the poorly understood M18 peptidases. DNPEP has implicated roles in protein and peptide metabolism, as well as the renin-angiotensin system in blood pressure regulation. Despite previous enzyme and substrate characterization, structural details of DNPEP regarding ligand recognition and catalytic mechanism remain to be delineated. Results: The crystal structure of human DNPEP complexed with zinc and a substrate analogue aspartate-β- hydroxamate reveals a dodecameric machinery built by domain-swapped dimers, in agreement with electron microscopy data. A structural comparison with bacterial homologues identifies unifying catalytic features among the poorly understood M18 enzymes. The bound ligands in the active site also reveal the coordination mode of the binuclear zinc centre and a substrate specificity pocket for acidic amino acids. Conclusions: The DNPEP structure provides a molecular framework to understand its catalysis that is mediated by active site loop swapping, a mechanism likely adopted in other M18 and M42 metallopeptidases that form dodecameric complexes as a self-compartmentalization strategy. Small differences in the substrate binding pocket such as shape and positive charges, the latter conferred by a basic lysine residue, further provide the key to distinguishing substrate preference. -

(12) United States Patent (10) Patent No.: US 8,603,824 B2 Ramseier Et Al
USOO8603824B2 (12) United States Patent (10) Patent No.: US 8,603,824 B2 Ramseier et al. (45) Date of Patent: Dec. 10, 2013 (54) PROCESS FOR IMPROVED PROTEIN 5,399,684 A 3, 1995 Davie et al. EXPRESSION BY STRAIN ENGINEERING 5,418, 155 A 5/1995 Cormier et al. 5,441,934 A 8/1995 Krapcho et al. (75) Inventors: Thomas M. Ramseier, Poway, CA 5,508,192 A * 4/1996 Georgiou et al. .......... 435/252.3 (US); Hongfan Jin, San Diego, CA 5,527,883 A 6/1996 Thompson et al. (US); Charles H. Squires, Poway, CA 5,558,862 A 9, 1996 Corbinet al. 5,559,015 A 9/1996 Capage et al. (US) 5,571,694 A 11/1996 Makoff et al. (73) Assignee: Pfenex, Inc., San Diego, CA (US) 5,595,898 A 1/1997 Robinson et al. 5,610,044 A 3, 1997 Lam et al. (*) Notice: Subject to any disclaimer, the term of this 5,621,074 A 4/1997 Bjorn et al. patent is extended or adjusted under 35 5,622,846 A 4/1997 Kiener et al. 5,641,671 A 6/1997 Bos et al. U.S.C. 154(b) by 471 days. 5,641,870 A 6/1997 Rinderknecht et al. 5,643,774 A 7/1997 Ligon et al. (21) Appl. No.: 11/189,375 5,662,898 A 9/1997 Ligon et al. (22) Filed: Jul. 26, 2005 5,677,127 A 10/1997 Hogan et al. 5,683,888 A 1 1/1997 Campbell (65) Prior Publication Data 5,686,282 A 11/1997 Lam et al. -

Serine Proteases with Altered Sensitivity to Activity-Modulating
(19) & (11) EP 2 045 321 A2 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 08.04.2009 Bulletin 2009/15 C12N 9/00 (2006.01) C12N 15/00 (2006.01) C12Q 1/37 (2006.01) (21) Application number: 09150549.5 (22) Date of filing: 26.05.2006 (84) Designated Contracting States: • Haupts, Ulrich AT BE BG CH CY CZ DE DK EE ES FI FR GB GR 51519 Odenthal (DE) HU IE IS IT LI LT LU LV MC NL PL PT RO SE SI • Coco, Wayne SK TR 50737 Köln (DE) •Tebbe, Jan (30) Priority: 27.05.2005 EP 05104543 50733 Köln (DE) • Votsmeier, Christian (62) Document number(s) of the earlier application(s) in 50259 Pulheim (DE) accordance with Art. 76 EPC: • Scheidig, Andreas 06763303.2 / 1 883 696 50823 Köln (DE) (71) Applicant: Direvo Biotech AG (74) Representative: von Kreisler Selting Werner 50829 Köln (DE) Patentanwälte P.O. Box 10 22 41 (72) Inventors: 50462 Köln (DE) • Koltermann, André 82057 Icking (DE) Remarks: • Kettling, Ulrich This application was filed on 14-01-2009 as a 81477 München (DE) divisional application to the application mentioned under INID code 62. (54) Serine proteases with altered sensitivity to activity-modulating substances (57) The present invention provides variants of ser- screening of the library in the presence of one or several ine proteases of the S1 class with altered sensitivity to activity-modulating substances, selection of variants with one or more activity-modulating substances. A method altered sensitivity to one or several activity-modulating for the generation of such proteases is disclosed, com- substances and isolation of those polynucleotide se- prising the provision of a protease library encoding poly- quences that encode for the selected variants. -

Association Between Chymase Gene Polymorphisms and Atrial Fibrillation
Zhou et al. BMC Cardiovascular Disorders (2019) 19:321 https://doi.org/10.1186/s12872-019-01300-7 RESEARCH ARTICLE Open Access Association between chymase gene polymorphisms and atrial fibrillation in Chinese Han population Dongchen Zhou, Yuewei Chen, Jiaxin Wu, Jiabo Shen, Yushan Shang, Liangrong Zheng and Xudong Xie* Abstract Background: Chymase is the major angiotensin II (Ang II)-forming enzyme in cardiovascular tissue, with an important role in atrial remodeling. This study aimed to examine the association between chymase 1 gene (CMA1) polymorphisms and atrial fibrillation (AF) in a Chinese Han population. Methods: This case-control study enrolled 126 patients with lone AF and 120 age- and sex-matched healthy controls, all from a Chinese Han population. Five CMA1 polymorphisms were genotyped. Results: The CMA1 polymorphism rs1800875 (G-1903A) was associated with AF. The frequency of the GG genotype was significantly higher in AF patients compared with controls (p = 0.009). Haplotype analysis further demonstrated an increased risk of AF associated with the rs1800875-G haplotype (Hap8 TGTTG, odds ratio (OR) = 1.668, 95% CI 1.132–2.458, p = 0.009), and a decreased risk for the rs1800875-A haplotype (Hap5 TATTG, OR = 0.178, 95% CI 0.042– 0.749, p = 0.008). Conclusions: CMA1 polymorphisms may be associated with AF, and the rs1800875 GG genotype might be a susceptibility factor for AF in the Chinese Han population. Keywords: Atrial fibrillation, CMA1, Chymase, Single nucleotide polymorphism, Angiotensin II Background ‘lone AF’, occur in the absence of identifiable underlying Atrial fibrillation (AF) is the most common type of sus- cardiovascular or other comorbid diseases [4]. -

CMA1 Rabbit Polyclonal Antibody – TA342809 | Origene
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for TA342809 CMA1 Rabbit Polyclonal Antibody Product data: Product Type: Primary Antibodies Applications: WB Recommended Dilution: WB Reactivity: Human Host: Rabbit Isotype: IgG Clonality: Polyclonal Immunogen: The immunogen for anti-CMA1 antibody: synthetic peptide directed towards the C terminal of human CMA1. Synthetic peptide located within the following region: EVKLRLMDPQACSHFRDFDHNLQLCVGNPRKTKSAFKGDSGGPLLCAGVA Formulation: Liquid. Purified antibody supplied in 1x PBS buffer with 0.09% (w/v) sodium azide and 2% sucrose. Note that this product is shipped as lyophilized powder to China customers. Conjugation: Unconjugated Storage: Store at -20°C as received. Stability: Stable for 12 months from date of receipt. Predicted Protein Size: 27 kDa Gene Name: chymase 1 Database Link: NP_001827 Entrez Gene 1215 Human P23946 Background: CMA1 is a chymotryptic serine proteinase that belongs to the peptidase family S1. It is expressed in mast cells and thought to function in the degradation of the extracellular matrix, the regulation of submucosal gland secretion, and the generation of vasoactive peptides. In the heart and blood vessels, this protein, rather than angiotensin converting enzyme, is largely responsible for converting angiotensin I to the vasoactive peptide angiotensin II. Angiotensin II has been implicated in blood pressure control and in the pathogenesis of hypertension, cardiac hypertrophy, and heart failure. Thus, this gene product is a target for cardiovascular disease therapies. This product is to be used for laboratory only. Not for diagnostic or therapeutic use. -

(12) Patent Application Publication (10) Pub. No.: US 2006/0110747 A1 Ramseier Et Al
US 200601 10747A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2006/0110747 A1 Ramseier et al. (43) Pub. Date: May 25, 2006 (54) PROCESS FOR IMPROVED PROTEIN (60) Provisional application No. 60/591489, filed on Jul. EXPRESSION BY STRAIN ENGINEERING 26, 2004. (75) Inventors: Thomas M. Ramseier, Poway, CA Publication Classification (US); Hongfan Jin, San Diego, CA (51) Int. Cl. (US); Charles H. Squires, Poway, CA CI2O I/68 (2006.01) (US) GOIN 33/53 (2006.01) CI2N 15/74 (2006.01) Correspondence Address: (52) U.S. Cl. ................................ 435/6: 435/7.1; 435/471 KING & SPALDING LLP 118O PEACHTREE STREET (57) ABSTRACT ATLANTA, GA 30309 (US) This invention is a process for improving the production levels of recombinant proteins or peptides or improving the (73) Assignee: Dow Global Technologies Inc., Midland, level of active recombinant proteins or peptides expressed in MI (US) host cells. The invention is a process of comparing two genetic profiles of a cell that expresses a recombinant (21) Appl. No.: 11/189,375 protein and modifying the cell to change the expression of a gene product that is upregulated in response to the recom (22) Filed: Jul. 26, 2005 binant protein expression. The process can improve protein production or can improve protein quality, for example, by Related U.S. Application Data increasing solubility of a recombinant protein. Patent Application Publication May 25, 2006 Sheet 1 of 15 US 2006/0110747 A1 Figure 1 09 010909070£020\,0 10°0 Patent Application Publication May 25, 2006 Sheet 2 of 15 US 2006/0110747 A1 Figure 2 Ester sers Custer || || || || || HH-I-H 1 H4 s a cisiers TT closers | | | | | | Ya S T RXFO 1961. -

1 No. Affymetrix ID Gene Symbol Genedescription Gotermsbp Q Value 1. 209351 at KRT14 Keratin 14 Structural Constituent of Cyto
1 Affymetrix Gene Q No. GeneDescription GOTermsBP ID Symbol value structural constituent of cytoskeleton, intermediate 1. 209351_at KRT14 keratin 14 filament, epidermis development <0.01 biological process unknown, S100 calcium binding calcium ion binding, cellular 2. 204268_at S100A2 protein A2 component unknown <0.01 regulation of progression through cell cycle, extracellular space, cytoplasm, cell proliferation, protein kinase C inhibitor activity, protein domain specific 3. 33323_r_at SFN stratifin/14-3-3σ binding <0.01 regulation of progression through cell cycle, extracellular space, cytoplasm, cell proliferation, protein kinase C inhibitor activity, protein domain specific 4. 33322_i_at SFN stratifin/14-3-3σ binding <0.01 structural constituent of cytoskeleton, intermediate 5. 201820_at KRT5 keratin 5 filament, epidermis development <0.01 structural constituent of cytoskeleton, intermediate 6. 209125_at KRT6A keratin 6A filament, ectoderm development <0.01 regulation of progression through cell cycle, extracellular space, cytoplasm, cell proliferation, protein kinase C inhibitor activity, protein domain specific 7. 209260_at SFN stratifin/14-3-3σ binding <0.01 structural constituent of cytoskeleton, intermediate 8. 213680_at KRT6B keratin 6B filament, ectoderm development <0.01 receptor activity, cytosol, integral to plasma membrane, cell surface receptor linked signal transduction, sensory perception, tumor-associated calcium visual perception, cell 9. 202286_s_at TACSTD2 signal transducer 2 proliferation, membrane <0.01 structural constituent of cytoskeleton, cytoskeleton, intermediate filament, cell-cell adherens junction, epidermis 10. 200606_at DSP desmoplakin development <0.01 lectin, galactoside- sugar binding, extracellular binding, soluble, 7 space, nucleus, apoptosis, 11. 206400_at LGALS7 (galectin 7) heterophilic cell adhesion <0.01 2 S100 calcium binding calcium ion binding, epidermis 12. 205916_at S100A7 protein A7 (psoriasin 1) development <0.01 S100 calcium binding protein A8 (calgranulin calcium ion binding, extracellular 13. -

Based on Network Pharmacology Tools to Investigate the Molecular Mechanism of Cordyceps Sinensis on the Treatment of Diabetic Nephropathy
Hindawi Journal of Diabetes Research Volume 2021, Article ID 8891093, 12 pages https://doi.org/10.1155/2021/8891093 Research Article Based on Network Pharmacology Tools to Investigate the Molecular Mechanism of Cordyceps sinensis on the Treatment of Diabetic Nephropathy Yan Li,1 Lei Wang,2 Bojun Xu,1 Liangbin Zhao,1 Li Li,1 Keyang Xu,3 Anqi Tang,1 Shasha Zhou,1 Lu Song,1 Xiao Zhang,1 and Huakui Zhan 1 1Hospital of Chengdu University of Traditional Chinese Medicine, Chengdu, 610072 Sichuan, China 2Key Laboratory of Chinese Internal Medicine of Ministry of Education and Dongzhimen Hospital, Beijing University of Chinese Medicine, Beijing 100700, China 3Zhejiang Chinese Medical University, Hangzhou, 310053 Zhejiang, China Correspondence should be addressed to Huakui Zhan; [email protected] Received 27 August 2020; Revised 17 January 2021; Accepted 24 January 2021; Published 8 February 2021 Academic Editor: Michaelangela Barbieri Copyright © 2021 Yan Li et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Background. Diabetic nephropathy (DN) is one of the most common complications of diabetes mellitus and is a major cause of end- stage kidney disease. Cordyceps sinensis (Cordyceps, Dong Chong Xia Cao) is a widely applied ingredient for treating patients with DN in China, while the molecular mechanisms remain unclear. This study is aimed at revealing the therapeutic mechanisms of Cordyceps in DN by undertaking a network pharmacology analysis. Materials and Methods. In this study, active ingredients and associated target proteins of Cordyceps sinensis were obtained via Traditional Chinese Medicine Systems Pharmacology Database (TCMSP) and Swiss Target Prediction platform, then reconfirmed by using PubChem databases. -
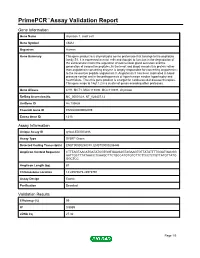
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name chymase 1, mast cell Gene Symbol CMA1 Organism Human Gene Summary This gene product is a chymotryptic serine proteinase that belongs to the peptidase family S1. It is expressed in mast cells and thought to function in the degradation of the extracellular matrix the regulation of submucosal gland secretion and the generation of vasoactive peptides. In the heart and blood vessels this protein rather than angiotensin converting enzyme is largely responsible for converting angiotensin I to the vasoactive peptide angiotensin II. Angiotensin II has been implicated in blood pressure control and in the pathogenesis of hypertension cardiac hypertrophy and heart failure. Thus this gene product is a target for cardiovascular disease therapies. This gene maps to 14q11.2 in a cluster of genes encoding other proteases. Gene Aliases CYH, MCT1, MGC119890, MGC119891, chymase RefSeq Accession No. NC_000014.8, NT_026437.12 UniGene ID Hs.135626 Ensembl Gene ID ENSG00000092009 Entrez Gene ID 1215 Assay Information Unique Assay ID qHsaCED0003486 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000250378, ENST00000206446 Amplicon Context Sequence CTTTAGTAACATGATATCGTGGTGAAGAGTAGAAGTGTTATATTTTGGATGACGG AATTGCTTTATAACCTCAAGCTTCTGCCATGTGTCTTCTTCCTCTGTTATGTTATG GGCTCC Amplicon Length (bp) 87 Chromosome Location 14:24975675-24975791 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 99 R2 0.9989 cDNA Cq 27.32 Page 1/5 PrimePCR™Assay Validation Report cDNA Tm (Celsius) 78 -

Human Induced Pluripotent Stem Cell–Derived Podocytes Mature Into Vascularized Glomeruli Upon Experimental Transplantation
BASIC RESEARCH www.jasn.org Human Induced Pluripotent Stem Cell–Derived Podocytes Mature into Vascularized Glomeruli upon Experimental Transplantation † Sazia Sharmin,* Atsuhiro Taguchi,* Yusuke Kaku,* Yasuhiro Yoshimura,* Tomoko Ohmori,* ‡ † ‡ Tetsushi Sakuma, Masashi Mukoyama, Takashi Yamamoto, Hidetake Kurihara,§ and | Ryuichi Nishinakamura* *Department of Kidney Development, Institute of Molecular Embryology and Genetics, and †Department of Nephrology, Faculty of Life Sciences, Kumamoto University, Kumamoto, Japan; ‡Department of Mathematical and Life Sciences, Graduate School of Science, Hiroshima University, Hiroshima, Japan; §Division of Anatomy, Juntendo University School of Medicine, Tokyo, Japan; and |Japan Science and Technology Agency, CREST, Kumamoto, Japan ABSTRACT Glomerular podocytes express proteins, such as nephrin, that constitute the slit diaphragm, thereby contributing to the filtration process in the kidney. Glomerular development has been analyzed mainly in mice, whereas analysis of human kidney development has been minimal because of limited access to embryonic kidneys. We previously reported the induction of three-dimensional primordial glomeruli from human induced pluripotent stem (iPS) cells. Here, using transcription activator–like effector nuclease-mediated homologous recombination, we generated human iPS cell lines that express green fluorescent protein (GFP) in the NPHS1 locus, which encodes nephrin, and we show that GFP expression facilitated accurate visualization of nephrin-positive podocyte formation in -

Gene Expression Profiling of Lymph Node Sub-Capsular Sinus Macrophages in Cancer
ORIGINAL RESEARCH published: 08 June 2021 doi: 10.3389/fimmu.2021.672123 Gene Expression Profiling of Lymph Node Sub-Capsular Sinus Macrophages in Cancer † † Danilo Pellin 1 , Natalie Claudio 2,3 , Zihan Guo 2,4, Tahereh Ziglari 2 and Ferdinando Pucci 2,3* 1 Gene Therapy Program, Dana-Farber/Boston Children’s Cancer and Blood Disorders Center, Harvard Medical School, Boston, MA, United States, 2 Department of Otolaryngology – Head and Neck Surgery, Oregon Health and Science University, Portland, OR, United States, 3 Department of Cell, Developmental & Cancer Biology, Oregon Health and Science University, Portland, OR, United States, 4 Program in Cancer Biology, Oregon Health and Science University, Portland, OR, United States Lymph nodes are key lymphoid organs collecting lymph fluid and migratory cells from the tissue area they survey. When cancerous cells arise within a tissue, the sentinel lymph node is the first immunological organ to mount an immune response. Sub-capsular sinus Edited by: Karine Rachel Prudent Breckpot, macrophages (SSMs) are specialized macrophages residing in the lymph nodes that play Vrije University Brussel, Belgium important roles as gatekeepers against particulate antigenic material. In the context of Reviewed by: cancer, SSMs capture tumor-derived extracellular vesicles (tEVs), a form of particulate Ioannis S. Pateras, National and Kapodistrian University of antigen released in high amounts by tumor cells. We and others have recently Athens, Greece demonstrated that SSMs possess anti-tumor activity because in their absence tumors Antonio Giovanni Solimando, progress faster. A comprehensive profiling of SSMs represents an important first step to University of Bari Aldo Moro, Italy Rohit Singh, identify the cellular and molecular mechanisms responsible for SSM anti-tumor activity.