Proposal for Bacillus Smithii Sp. Nov
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Heterotrophic Denitrification by Gram-Positive Bacteria: Bacillus Cereus and Bacillus Tequilensis
International Journal of Scientific and Research Publications, Volume 4, Issue 4, April 2014 1 ISSN 2250-3153 Heterotrophic denitrification by Gram-positive bacteria: Bacillus cereus and Bacillus tequilensis Moukhlissi Saïd*, Aboussabiq Fatima Ezzahra*, Amine Jamal*, Rihani Mohammed* and Assobhei Omar* * Laboratory of Marine Biotechnology and Environment, Faculty of Science of El Jadida, P.O. Box 20, El Jadida 24000, Morocco. Abstract- Two bacteria were isolated from anoxic denitrifying notoriously overlooked in community analysis of denitrifiers in reactor for treatment of domestic wastewater. The analysis of the the environment because they are not targeted by the available 16S rDNA gene sequences showed that the isolated strains were PCR primers designed for denitrification genes (Throbäck et al., affiliated with Bacillus cereus and Bacillus tequilensis. 2004). Verbaendert et al. (2011) have studied the denitrification Denitrification was compared between Bacillus cereus and of a large collection of Bacillus strains and suggested that Bacillus tequilensis in this study. Two bacilli were able to denitrification occurred in nearly half of the analysed strains. denitrify and Bacillus cereus was more efficient than Bacillus More recently, a variety of bacilli were tested for gas production tequilensis. Bacillus cereus reduced 80% of high amount of under denitrifying conditions and found to be complete nitrate; however, Bacillus tequilensis could reduce 37.4% of denitrifiers (Jones et al., 2011). Genome sequencing has revealed nitrate. These heterotrophic bacteria are able to eliminate organic the potential for partial denitrification in some Bacillus species. matter with the same trend reducing 74.5% for Bacillus For example, qNor is present in Bacillus tusciae strain DSM2912 tequilensis and 70.2% for Bacillus cereus. -

Food Waste Composting and Microbial Community Structure Profiling
processes Review Food Waste Composting and Microbial Community Structure Profiling Kishneth Palaniveloo 1,* , Muhammad Azri Amran 1, Nur Azeyanti Norhashim 1 , Nuradilla Mohamad-Fauzi 1,2, Fang Peng-Hui 3, Low Hui-Wen 3, Yap Kai-Lin 3, Looi Jiale 3, Melissa Goh Chian-Yee 3, Lai Jing-Yi 3, Baskaran Gunasekaran 3,* and Shariza Abdul Razak 4,* 1 Institute of Ocean and Earth Sciences, Institute for Advanced Studies Building, University of Malaya, Wilayah Persekutuan Kuala Lumpur 50603, Malaysia; [email protected] (M.A.A.); [email protected] (N.A.N.) 2 Institute of Biological Sciences, Faculty of Science, University of Malaya, Wilayah Persekutuan Kuala Lumpur 50603, Malaysia; [email protected] 3 Faculty of Applied Science, UCSI University (South Wing), Cheras, Wilayah Persekutuan Kuala Lumpur 56000, Malaysia; [email protected] (F.P.-H.); [email protected] (L.H.-W.); [email protected] (Y.K.-L.); [email protected] (L.J.); [email protected] (M.G.C.-Y.); [email protected] (L.J.-Y.) 4 Nutrition and Dietetics Program, School of Health Sciences, Health Campus, Universiti Sains Malaysia, Kubang Kerian 16150, Kelantan, Malaysia * Correspondence: [email protected] (K.P.); [email protected] (B.G.); [email protected] (S.A.R.); Tel.: +60-3-7967-4640 (K.P.); +60-16-323-4159 (B.G.); +60-19-964-4043 (S.A.R.) Received: 20 May 2020; Accepted: 16 June 2020; Published: 22 June 2020 Abstract: Over the last decade, food waste has been one of the major issues globally as it brings a negative impact on the environment and health. -

Mai Muun Muntant Un an to the Man Uniti
MAIMUUN MUNTANTUS009855303B2 UN AN TO THEMAN UNITI (12 ) United States Patent ( 10 ) Patent No. : US 9 , 855 ,303 B2 McKenzie et al. (45 ) Date of Patent: * Jan . 2 , 2018 ( 54 ) COMPOSITIONS AND METHODS (58 ) Field of Classification Search ??? . A61K 35 / 742 (71 ) Applicant : SERES THERAPEUTICS , INC . , See application file for complete search history . Cambridge, MA (US ) (72 ) Inventors : Gregory McKenzie , Arlington , MA ( 56 ) References Cited (US ) ; Mary - Jane Lombardo McKenzie , Arlington , MA (US ) ; David U . S . PATENT DOCUMENTS N . Cook , Brooklyn , NY (US ) ; Marin 3 ,009 , 864 A 11 / 1961 Gordon - Aldterton et al. Vulic , Boston , MA (US ) ; Geoffrey von 3 ,228 , 838 A 1 / 1966 Rinfret Maltzahn , Boston , MA (US ) ; Brian 3 ,608 ,030 A 9 / 1971 Tint Goodman , Boston , MA (US ) ; John 4 ,077 , 227 A 3 / 1978 Larson Grant Aunins , Doylestown , PA (US ) ; 4 , 205 , 132 A 5 / 1980 Sandine Matthew R . Henn , Somerville , MA 4 ,655 ,047 A 4 / 1987 Temple (US ) ; David Arthur Berry , Brookline , 4 ,689 , 226 A 8 / 1987 Nurmi MA (US ) ; Jonathan Winkler , Boston , 4 ,839 , 281 A 6 / 1989 Gorbach et al . 5 , 196 , 205 A 3 / 1993 Borody MA (US ) 5 , 425 , 951 A 6 / 1995 Goodrich 5 ,436 ,002 A 7 / 1995 Payne ( 73 ) Assignee : Seres Therapeutics , Inc ., Cambridge , 5 ,443 ,826 A 8 / 1995 Borody MA (US ) 5 , 599 , 795 A 2 / 1997 McCann 5 ,648 ,206 A 7 / 1997 Goodrich ( * ) Notice : Subject to any disclaimer , the term of this 5 , 951 , 977 A 9 / 1999 Nisbet et al. patent is extended or adjusted under 35 5 , 965 , 128 A 10 / 1999 Doyle et al . -

WO 2016/086210 Al 2 June 2016 (02.06.2016) P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2016/086210 Al 2 June 2016 (02.06.2016) P O P C T (51) International Patent Classification: (74) Agents: MELLO, Jill, Ann et al; McCarter & English, A61P 1/00 (2006.01) A61P 37/00 (2006.01) LLP, 265 Franklin Street, Boston, MA 021 10 (US). A61P 29/00 (2006.01) A61K 35/74 (2015.01) (81) Designated States (unless otherwise indicated, for every (21) International Application Number: kind of national protection available): AE, AG, AL, AM, PCT/US20 15/0628 10 AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, (22) International Filing Date: DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, 25 November 2015 (25.1 1.2015) HN, HR, HU, ID, IL, IN, IR, IS, JP, KE, KG, KN, KP, KR, (25) Filing Language: English KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM, (26) Publication Language: English PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, SC, (30) Priority Data: SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, 62/084,537 25 November 2014 (25. 11.2014) US TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. 62/084,536 25 November 2014 (25. -

Manganese-II Oxidation and Copper-II Resistance in Endospore Forming Firmicutes Isolated from Uncontaminated Environmental Sites
AIMS Environmental Science, 3(2): 220-238. DOI: 10.3934/environsci.2016.2.220 Received 21 January 2016, Accepted 05 April 2016, Published 12 April 2016 http://www.aimspress.com/journal/environmental Research article Manganese-II oxidation and Copper-II resistance in endospore forming Firmicutes isolated from uncontaminated environmental sites Ganesan Sathiyanarayanan 1,†, Sevasti Filippidou 1,†, Thomas Junier 1,2, Patricio Muñoz Rufatt 1, Nicole Jeanneret 1, Tina Wunderlin 1, Nathalie Sieber 1, Cristina Dorador 3 and Pilar Junier 1,* 1 Laboratory of Microbiology, Institute of Biology, University of Neuchâtel, Emile-Argand 11, CH-2000 Neuchâtel, Switzerland 2 Vital-IT group, Swiss Institute of Bioinformatics, CH-1000 Lausanne, Switzerland 3 Laboratorio de Complejidad Microbiana y Ecología Funcional; Departamento de Biotecnología; Facultad de Ciencias del Mar y Recursos Biológicos, Universidad de Antofagasta; CL-, 1270190, Antofagasta, Chile * Correspondence: Email: [email protected]; Tel: +41-32-718-2244; Fax: +41-32-718-3001. † These authors contributed equally to this work. Abstract: The accumulation of metals in natural environments is a growing concern of modern societies since they constitute persistent, non-degradable contaminants. Microorganisms are involved in redox processes and participate to the biogeochemical cycling of metals. Some endospore-forming Firmicutes (EFF) are known to oxidize and reduce specific metals and have been isolated from metal-contaminated sites. However, whether EFF isolated from uncontaminated sites have the same capabilities has not been thoroughly studied. In this study, we measured manganese oxidation and copper resistance of aerobic EFF from uncontaminated sites. For the purposes of this study we have sampled 22 natural habitats and isolated 109 EFF strains. -

Identification and Characterization of Bacillus Cereus
Journal of Insect Science RESEARCH Identification and Characterization of Bacillus cereus SW7-1 in Bombyx mori (Lepidoptera: Bombycidae) Guan-Nan Li,1,2 Xue-Juan Xia,3 Huan-Huan Zhao,1,2 Parfait Sendegeya,1,2 and Yong Zhu1,2,4 1College of Biotechnology, Southwest University, Chongqing 400716, China 2State Key Laboratory of Silkworm Genome Biology, Chongqing 400716, China 3College of Food Science, Southwest University, Chongqing 400716, China 4Corresponding author, e-mail: [email protected] Subject Editor: Seth Barribeau J. Insect Sci. (2015) 15(1): 136; DOI: 10.1093/jisesa/iev121 ABSTRACT. The bacterial diseases of silkworms cause significant reductions in sericulture and result in huge economic loss. This study aimed to identify and characterize a pathogen from diseased silkworm. SW7-1, a pathogenic bacterial strain, was isolated from the dis- eased silkworm. The strain was identified on the basis of its bacteriological properties and 16S rRNA gene sequence. The colony was round, slightly convex, opaque, dry, and milky on a nutrient agar medium, the colony also exhibited jagged edges. SW7-1 was Gram- positive, without parasporal crystal, and 0.8–1.2 by 2.6–3.4 mm in length, resembling long rods with rounded ends. The strain was posi- tive to most of the physiological biochemical tests used in this study. The strain could utilize glucose, sucrose, and maltose. The results of its 16S rRNA gene sequence analysis revealed that SW7-1 shared the highest sequence identity (>99%) with Bacillus cereus strain 14. The bacterial strain was highly susceptible to gentamycin, streptomycin, erythromycin, norfloxacin, and ofloxacin and moderately susceptible to tetracycline and rifampicin. -

Diversity and Biocontrol Potential of Cultivable Endophytic Bacteria Associated with Halophytes from the West Aral Sea Basin
microorganisms Article Diversity and Biocontrol Potential of Cultivable Endophytic Bacteria Associated with Halophytes from the West Aral Sea Basin Lei Gao 1,2, Jinbiao Ma 1 , Yonghong Liu 1 , Yin Huang 1, Osama Abdalla Abdelshafy Mohamad 1 , Hongchen Jiang 1,3 , Dilfuza Egamberdieva 4,5 , Wenjun Li 1,6,* and Li Li 1,* 1 State Key Laboratory of Desert and Oasis Ecology, Xinjiang Institute of Ecology and Geography, Chinese Academy of Sciences, Urumqi 830011, China; [email protected] (L.G.); [email protected] (J.M.); [email protected] (Y.L.); [email protected] (Y.H.); [email protected] (O.A.A.M.); [email protected] (H.J.) 2 College of Resources and Environment, University of Chinese Academy of Sciences, Beijing 100049, China 3 State Key Laboratory of Biogeology and Environmental Geology, China University of Geosciences, Wuhan 430074, China 4 Faculty of Biology, National University of Uzbekistan, Tashkent 100174, Uzbekistan; [email protected] 5 Leibniz Centre for Agricultural Landscape Research (ZALF), 15374 Müncheberg, Germany 6 State Key Laboratory of Biocontrol, Guangdong Provincial Key Laboratory of Plant Resources, School of Life Sciences, Sun Yat-Sen University, Guangzhou 510275, China * Correspondence: [email protected] (W.L.); [email protected] (L.L.) Abstract: Endophytes associated with halophytes may contribute to the host’s adaptation to adverse Citation: Gao, L.; Ma, J.; Liu, Y.; environmental conditions through improving their stress tolerance and protecting them from various Huang, Y.; Mohamad, O.A.A.; Jiang, soil-borne pathogens. In this study, the diversity and antifungal activity of endophytic bacteria H.; Egamberdieva, D.; Li, W.; Li, L. -
![Downloaded from the Microbial Genome (Complete and Non-Classical Protein Secretion [73]](https://docslib.b-cdn.net/cover/5140/downloaded-from-the-microbial-genome-complete-and-non-classical-protein-secretion-73-3625140.webp)
Downloaded from the Microbial Genome (Complete and Non-Classical Protein Secretion [73]
Helen et al. BMC Genomics (2016) 17:155 DOI 10.1186/s12864-016-2465-0 RESEARCH ARTICLE Open Access Highly diverse nirK genes comprise two major clades that harbour ammonium- producing denitrifiers Decleyre Helen, Heylen Kim*, Bjorn Tytgat and Willems Anne* Abstract Background: Copper dependent nitrite reductase, NirK, catalyses the key step in denitrification, i.e. nitrite reduction to nitric oxide. Distinct structural NirK classes and phylogenetic clades of NirK-type denitrifiers have previously been observed based on a limited set of NirK sequences, however, their environmental distribution or ecological strategies are currently unknown. In addition, environmental nirK-type denitrifiers are currently underestimated in PCR-dependent surveys due to primer coverage limitations that can be attributed to their broad taxonomic diversity and enormous nirK sequence divergence. Therefore, we revisited reported analyses on partial NirK sequences using a taxonomically diverse, full-length NirK sequence dataset. Results: Division of NirK sequences into two phylogenetically distinct clades was confirmed, with Clade I mainly comprising Alphaproteobacteria (plus some Gamma- and Betaproteobacteria) and Clade II harbouring more diverse taxonomic groups like Archaea, Bacteroidetes, Chloroflexi, Gemmatimonadetes, Nitrospirae, Firmicutes, Actinobacteria, Planctomycetes and Proteobacteria (mainly Beta and Gamma). Failure of currently available primer sets to target diverse NirK-type denitrifiers in environmental surveys could be attributed to mismatches over the whole length of the primer binding regions including the 3′ site, with Clade II sequences containing higher sequence divergence than Clade I sequences. Simultaneous presence of both the denitrification and DNRA pathway could be observed in 67 % of all NirK-type denitrifiers. Conclusion: The previously reported division of NirK into two distinct phylogenetic clades was confirmed using a taxonomically diverse set of full-length NirK sequences. -

Microbial Hitchhikers on Intercontinental Dust: Catching a Lift in Chad
The ISME Journal (2013) 7, 850–867 & 2013 International Society for Microbial Ecology All rights reserved 1751-7362/13 www.nature.com/ismej ORIGINAL ARTICLE Microbial hitchhikers on intercontinental dust: catching a lift in Chad Jocelyne Favet1, Ales Lapanje2, Adriana Giongo3, Suzanne Kennedy4, Yin-Yin Aung1, Arlette Cattaneo1, Austin G Davis-Richardson3, Christopher T Brown3, Renate Kort5, Hans-Ju¨ rgen Brumsack6, Bernhard Schnetger6, Adrian Chappell7, Jaap Kroijenga8, Andreas Beck9,10, Karin Schwibbert11, Ahmed H Mohamed12, Timothy Kirchner12, Patricia Dorr de Quadros3, Eric W Triplett3, William J Broughton1,11 and Anna A Gorbushina1,11,13 1Universite´ de Gene`ve, Sciences III, Gene`ve 4, Switzerland; 2Institute of Physical Biology, Ljubljana, Slovenia; 3Department of Microbiology and Cell Science, Institute of Food and Agricultural Sciences, University of Florida, Gainesville, FL, USA; 4MO BIO Laboratories Inc., Carlsbad, CA, USA; 5Elektronenmikroskopie, Carl von Ossietzky Universita¨t, Oldenburg, Germany; 6Microbiogeochemie, ICBM, Carl von Ossietzky Universita¨t, Oldenburg, Germany; 7CSIRO Land and Water, Black Mountain Laboratories, Black Mountain, ACT, Australia; 8Konvintsdyk 1, Friesland, The Netherlands; 9Botanische Staatssammlung Mu¨nchen, Department of Lichenology and Bryology, Mu¨nchen, Germany; 10GeoBio-Center, Ludwig-Maximilians Universita¨t Mu¨nchen, Mu¨nchen, Germany; 11Bundesanstalt fu¨r Materialforschung, und -pru¨fung, Abteilung Material und Umwelt, Berlin, Germany; 12Geomatics SFRC IFAS, University of Florida, Gainesville, FL, USA and 13Freie Universita¨t Berlin, Fachbereich Biologie, Chemie und Pharmazie & Geowissenschaften, Berlin, Germany Ancient mariners knew that dust whipped up from deserts by strong winds travelled long distances, including over oceans. Satellite remote sensing revealed major dust sources across the Sahara. Indeed, the Bode´le´ Depression in the Republic of Chad has been called the dustiest place on earth. -

Draft Genome Sequence of Bacillus Azotoformans MEV2011, a (Co
Nielsen et al. Standards in Genomic Sciences 2015, 9:23 http://www.standardsingenomics.com/content/9/1/23 SHORT GENOME REPORT Open Access Draft genome sequence of Bacillus azotoformans MEV2011, a (Co-) denitrifying strain unable to grow with oxygen Maja Nielsen1†, Lars Schreiber2†, Kai Finster1,3 and Andreas Schramm1,2* Abstract Bacillus azotoformans MEV2011, isolated from soil, is a microaerotolerant obligate denitrifier, which can also produce N2 by co-denitrification. Oxygen is consumed but not growth-supportive. The draft genome has a size of 4.7 Mb and contains key genes for both denitrification and dissimilatory nitrate reduction to ammonium. Keywords: Bacillus azotoformans, Denitrification, Codenitrification, Oxygen Introduction 15N incubations; data not shown) starts at microaerobic Species of the genus Bacillus are characterized as Gram- conditions (<30 μMO2; Figure 2), yet the initial pres- positive, facultative aerobic bacteria capable of forming ence of oxygen in the growth medium leads to longer endospores [1]. In the absence of oxygen, many Bacillus lag phases and no increase in final density of the culture species can respire with nitrate instead, employing either (Figure 3); growth without nitrate was never observed. dissimilatory nitrate reduction to ammonium or denitrifi- Therefore, we characterize B. azotoformans MEV2011 cation [2,3]. Despite the widespread occurrence of nitrate- as microaerotolerant obligate denitrifier. In addition, reducing bacilli, their molecular and genetic basis remained B. azotoformans MEV2011 is capable of co-denitrification, poorly investigated [4,5]. Only recently, genome sequencing a co-metabolic process, in which reduced nitrogen com- of two denitrifying type strains, B. azotoformans LMG 9581T pounds like amino acids or hydroxylamine react with NO+ T and B. -

Bacillus Coagulans SNZ1969 Spores Preparation
GRAS Notice (GRN) No. 597 GR 11111111 111111 II 1111 http://www.fda.gov/Food/IngredientsPackagingLabeling/GRAS/NoticeInventory/default.htm ORIGINAL SUBMISSION oooooi 749 46th Square Vera Beach, FL 32968, USA Telephone: 772-299-0746 ~ont & ~ssoclates 3Jnc. Facsimile: 772-299-5381 E-mail: [email protected] ' '\'l August 24, 2015 Office of Food Additive Safety (HFS-255) Center for Food Safety and Applied Nutrition Food and Drug Administration 5100 Paint Branch Parkway College Park, MD 20740-3835 Subject: GRAS Notification for Bacillus coagulans SNZ1969 spore preparation Dear Sir/Madam: Pursuant to proposed 21 CFR 170.36 (62 FR 18960; April 17, 1997), Sanzyme Limited, India, through Soni & Associates Inc. as its agent, hereby provides notice of a claim that the food ingredient Bacillus coagulans SNZ1969 spores preparation described in the enclosed notification document is exempt from the premarket approval requirement of the Federal Food, Drug, and Cosmetic Act because it has been determined to be Generally Recognized As Safe (GRAS), based on scientific procedures. As required, please find enclosed three copies of the notification. If you have any questions or require additional information, please let me know by phone at 772-299-0746 or by email at msoni@ soniassociates.net or sonim@ bellsouth.net. (b) (6) Madhu G. Soni, PhD, FACN, FATS Agent for Sanzyme Limited INDIA 000002 AUG 2 6 .,.Q;) www.soniassociates.net OFFICE Or FOOD ADDITIVE SAFETY 749 46th Square Vera Beach, FL 32968, USA Telephone: 772-299-0746 ~out & ~~~octate~ Jfnc. Facsimile: 772-299-5381 E-mail: [email protected] www.soniassociates.net GRAS NOTIFICATION I. -
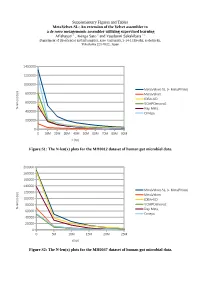
Supplementary Figures and Tables Metavelvet-SL
Supplementary Figures and Tables MetaVelvet-SL: An extension of the Velvet assembler to a de novo metagenomic assembler utilizing supervised learning Afiahayati 1 , Kengo Sato 1 and Yasubumi Sakakibara 1∗ Department of Biosciences and Informatics, Keio University, 3-14-1 Hiyoshi, Kohoku-ku, Yokohama 223-8522, Japan 1400000 1200000 1000000 MetaVelvet-SL (+ MetaPhlAn) ) p 800000 b MetaVelvet ( ) x ( IDBA-UD n 600000 e SOAPDenovo2 l - N Ray Meta 400000 Omega 200000 0 0 10M 20M 30M 40M 50M 60M 70M 80M 90M x (bp) Figure S1: The N-len(x) plots for the MH0012 dataset of human gut microbial data. 200000 180000 160000 140000 MetaVelvet-SL (+ MetaPhlAn) ) 120000 p b MetaVelvet ( ) 100000 x IDBA-UD ( n e l 80000 SOAPDenovo2 - N Ray Meta 60000 Omega 40000 20000 0 0 5M 10M 15M 20M 25M x(bp) Figure S2: The N-len(x) plots for the MH0047 dataset of human gut microbial data. 600000 500000 400000 MetaVelvet-SL (+ MetaPhlAn) ) p b MetaVelvet ( ) 300000 x IDBA-UD ( n e l SOAPDenovo2 - N 200000 Ray Meta Omega 100000 0 0 10M 20M 30M 40M 50M 60M 70M 80M 90M x (bp) Figure S3: The N-len(x) plots for the SRS017227 dataset of human gut microbial data. 800000 700000 600000 500000 MetaVelvet-SL (+ MetaPhlAn) ) p b MetaVelvet ( ) 400000 x IDBA-UD ( n e l SOAPDenovo2 - 300000 N Ray Meta 200000 Omega 100000 0 0 5M 10M 15M 20M 25M 30M 35M 40M 45M x (bp) Figure S4: The N-len(x) plots for the SRS018661 dataset of human gut microbial data. Formula to identify unique nodes in Velvet (Zerbino and Birney, 2008) ̄x2 ρ2− log2 2 F ( ̄x ,n,ρ )= +n 2 2 where ̄x = the coverage of node ρ = expected coverage of subgraph n = the length of node A node is “unique”, if its F > 5 .