Research Collection
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Allele-Specific Expression of Ribosomal Protein Genes in Interspecific Hybrid Catfish
Allele-specific Expression of Ribosomal Protein Genes in Interspecific Hybrid Catfish by Ailu Chen A dissertation submitted to the Graduate Faculty of Auburn University in partial fulfillment of the requirements for the Degree of Doctor of Philosophy Auburn, Alabama August 1, 2015 Keywords: catfish, interspecific hybrids, allele-specific expression, ribosomal protein Copyright 2015 by Ailu Chen Approved by Zhanjiang Liu, Chair, Professor, School of Fisheries, Aquaculture and Aquatic Sciences Nannan Liu, Professor, Entomology and Plant Pathology Eric Peatman, Associate Professor, School of Fisheries, Aquaculture and Aquatic Sciences Aaron M. Rashotte, Associate Professor, Biological Sciences Abstract Interspecific hybridization results in a vast reservoir of allelic variations, which may potentially contribute to phenotypical enhancement in the hybrids. Whether the allelic variations are related to the downstream phenotypic differences of interspecific hybrid is still an open question. The recently developed genome-wide allele-specific approaches that harness high- throughput sequencing technology allow direct quantification of allelic variations and gene expression patterns. In this work, I investigated allele-specific expression (ASE) pattern using RNA-Seq datasets generated from interspecific catfish hybrids. The objective of the study is to determine the ASE genes and pathways in which they are involved. Specifically, my study investigated ASE-SNPs, ASE-genes, parent-of-origins of ASE allele and how ASE would possibly contribute to heterosis. My data showed that ASE was operating in the interspecific catfish system. Of the 66,251 and 177,841 SNPs identified from the datasets of the liver and gill, 5,420 (8.2%) and 13,390 (7.5%) SNPs were identified as significant ASE-SNPs, respectively. -

CD8 T Cell Memory Transition and Maintenance of Functional Foxo1
FoxO1 Controls Effector-to-Memory Transition and Maintenance of Functional CD8 T Cell Memory This information is current as Melba Marie Tejera, Eui Ho Kim, Jeremy A. Sullivan, Erin of October 1, 2021. H. Plisch and M. Suresh J Immunol 2013; 191:187-199; Prepublished online 3 June 2013; doi: 10.4049/jimmunol.1300331 http://www.jimmunol.org/content/191/1/187 Downloaded from Supplementary http://www.jimmunol.org/content/suppl/2013/06/03/jimmunol.130033 Material 1.DC1 http://www.jimmunol.org/ References This article cites 52 articles, 7 of which you can access for free at: http://www.jimmunol.org/content/191/1/187.full#ref-list-1 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists by guest on October 1, 2021 • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2013 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology FoxO1 Controls Effector-to-Memory Transition and Maintenance of Functional CD8 T Cell Memory Melba Marie Tejera, Eui Ho Kim, Jeremy A. -

Supplementary Figures 1-14 and Supplementary References
SUPPORTING INFORMATION Spatial Cross-Talk Between Oxidative Stress and DNA Replication in Human Fibroblasts Marko Radulovic,1,2 Noor O Baqader,1 Kai Stoeber,3† and Jasminka Godovac-Zimmermann1* 1Division of Medicine, University College London, Center for Nephrology, Royal Free Campus, Rowland Hill Street, London, NW3 2PF, UK. 2Insitute of Oncology and Radiology, Pasterova 14, 11000 Belgrade, Serbia 3Research Department of Pathology and UCL Cancer Institute, Rockefeller Building, University College London, University Street, London WC1E 6JJ, UK †Present Address: Shionogi Europe, 33 Kingsway, Holborn, London WC2B 6UF, UK TABLE OF CONTENTS 1. Supplementary Figures 1-14 and Supplementary References. Figure S-1. Network and joint spatial razor plot for 18 enzymes of glycolysis and the pentose phosphate shunt. Figure S-2. Correlation of SILAC ratios between OXS and OAC for proteins assigned to the SAME class. Figure S-3. Overlap matrix (r = 1) for groups of CORUM complexes containing 19 proteins of the 49-set. Figure S-4. Joint spatial razor plots for the Nop56p complex and FIB-associated complex involved in ribosome biogenesis. Figure S-5. Analysis of the response of emerin nuclear envelope complexes to OXS and OAC. Figure S-6. Joint spatial razor plots for the CCT protein folding complex, ATP synthase and V-Type ATPase. Figure S-7. Joint spatial razor plots showing changes in subcellular abundance and compartmental distribution for proteins annotated by GO to nucleocytoplasmic transport (GO:0006913). Figure S-8. Joint spatial razor plots showing changes in subcellular abundance and compartmental distribution for proteins annotated to endocytosis (GO:0006897). Figure S-9. Joint spatial razor plots for 401-set proteins annotated by GO to small GTPase mediated signal transduction (GO:0007264) and/or GTPase activity (GO:0003924). -

1 AGING Supplementary Table 2
SUPPLEMENTARY TABLES Supplementary Table 1. Details of the eight domain chains of KIAA0101. Serial IDENTITY MAX IN COMP- INTERFACE ID POSITION RESOLUTION EXPERIMENT TYPE number START STOP SCORE IDENTITY LEX WITH CAVITY A 4D2G_D 52 - 69 52 69 100 100 2.65 Å PCNA X-RAY DIFFRACTION √ B 4D2G_E 52 - 69 52 69 100 100 2.65 Å PCNA X-RAY DIFFRACTION √ C 6EHT_D 52 - 71 52 71 100 100 3.2Å PCNA X-RAY DIFFRACTION √ D 6EHT_E 52 - 71 52 71 100 100 3.2Å PCNA X-RAY DIFFRACTION √ E 6GWS_D 41-72 41 72 100 100 3.2Å PCNA X-RAY DIFFRACTION √ F 6GWS_E 41-72 41 72 100 100 2.9Å PCNA X-RAY DIFFRACTION √ G 6GWS_F 41-72 41 72 100 100 2.9Å PCNA X-RAY DIFFRACTION √ H 6IIW_B 2-11 2 11 100 100 1.699Å UHRF1 X-RAY DIFFRACTION √ www.aging-us.com 1 AGING Supplementary Table 2. Significantly enriched gene ontology (GO) annotations (cellular components) of KIAA0101 in lung adenocarcinoma (LinkedOmics). Leading Description FDR Leading Edge Gene EdgeNum RAD51, SPC25, CCNB1, BIRC5, NCAPG, ZWINT, MAD2L1, SKA3, NUF2, BUB1B, CENPA, SKA1, AURKB, NEK2, CENPW, HJURP, NDC80, CDCA5, NCAPH, BUB1, ZWILCH, CENPK, KIF2C, AURKA, CENPN, TOP2A, CENPM, PLK1, ERCC6L, CDT1, CHEK1, SPAG5, CENPH, condensed 66 0 SPC24, NUP37, BLM, CENPE, BUB3, CDK2, FANCD2, CENPO, CENPF, BRCA1, DSN1, chromosome MKI67, NCAPG2, H2AFX, HMGB2, SUV39H1, CBX3, TUBG1, KNTC1, PPP1CC, SMC2, BANF1, NCAPD2, SKA2, NUP107, BRCA2, NUP85, ITGB3BP, SYCE2, TOPBP1, DMC1, SMC4, INCENP. RAD51, OIP5, CDK1, SPC25, CCNB1, BIRC5, NCAPG, ZWINT, MAD2L1, SKA3, NUF2, BUB1B, CENPA, SKA1, AURKB, NEK2, ESCO2, CENPW, HJURP, TTK, NDC80, CDCA5, BUB1, ZWILCH, CENPK, KIF2C, AURKA, DSCC1, CENPN, CDCA8, CENPM, PLK1, MCM6, ERCC6L, CDT1, HELLS, CHEK1, SPAG5, CENPH, PCNA, SPC24, CENPI, NUP37, FEN1, chromosomal 94 0 CENPL, BLM, KIF18A, CENPE, MCM4, BUB3, SUV39H2, MCM2, CDK2, PIF1, DNA2, region CENPO, CENPF, CHEK2, DSN1, H2AFX, MCM7, SUV39H1, MTBP, CBX3, RECQL4, KNTC1, PPP1CC, CENPP, CENPQ, PTGES3, NCAPD2, DYNLL1, SKA2, HAT1, NUP107, MCM5, MCM3, MSH2, BRCA2, NUP85, SSB, ITGB3BP, DMC1, INCENP, THOC3, XPO1, APEX1, XRCC5, KIF22, DCLRE1A, SEH1L, XRCC3, NSMCE2, RAD21. -

GIGSEA: Genotype Imputed Gene Set Enrichment Analysis
GIGSEA: Genotype Imputed Gene Set Enrichment Analysis Shijia Zhu Department of Genetics and Genomic Sciences and Icahn Institute for Genomics and Multiscale Biology, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA August 28, 2017 Contents Abstract . .1 1. Import packages . .1 2. Quick start . .2 3. One example of MetaXcan output . .2 4. Load data of gene sets . .4 4.1 Discrete-valued gene sets: . .4 4.2 Continuous-valued gene sets: . .6 4.3 User self-defined gene set . .7 5. Gene set enrichment analysis . .9 5.1 Gene set enrichment analysis using weighted simple linear regression . .9 5.2 Gene set enrichment analysis using weighted multiple regression model . 10 5.3 One-step weightedGSEA . 11 Abstract We presented the Genotype Imputed Gene Set Enrichment Analysis (GIGSEA), a novel method that uses GWAS-and-eQTL-imputed differential gene expression to interrogate gene set enrichment for the trait-associated SNPs. By incorporating eQTL from large gene expression studies, e.g. GTEx, GIGSEA appropriately addresses such challenges for SNP enrichment as gene size, gene boundary, andmultiple-marker regulation. The weighted linear regression model, taking as weights both imputation accuracy and model completeness, was used to test the enrichment, properly adjusting the bias due to redundancy in different gene sets. The permutation test, furthermore, is used to evaluate the significance of enrichment, whose efficiency can be largely elevated by expressing the computational intensive part in terms of large matrix operation. We have shown the appropriate type I error rates for GIGSEA (<5%), and the preliminary results also demonstrate its good performance to uncover the real biological signal. -

MRPL24 Polyclonal Antibody
PRODUCT DATA SHEET Bioworld Technology,Inc. MRPL24 polyclonal antibody Catalog: BS65388 Host: Rabbit Reactivity: Human,Mouse,Ra BackGround: term. Avoid freeze-thaw cycles. mitochondrial ribosomal protein L24(MRPL24) Homo Specificity: sapiens Mammalian mitochondrial ribosomal proteins are MRP-L24 Polyclonal Antibody detects endogenous levels encoded by nuclear genes and help in protein synthesis of MRP-L24 protein. within the mitochondrion. Mitochondrial ribosomes (mi- DATA: toribosomes) consist of a small 28S subunit and a large 39S subunit. They have an estimated 75% protein to rRNA composition compared to prokaryotic ribosomes, where this ratio is reversed. Another difference between mammalian mitoribosomes and prokaryotic ribosomes is that the latter contain a 5S rRNA. Among different spe- cies, the proteins comprising the mitoribosome differ greatly in sequence, and sometimes in biochemical prop- Western Blot analysis of various cells using MRP-L24 Polyclonal Anti- erties, which prevents easy recognition by sequence ho- body diluted at 1:2000 mology. This gene encodes a 39S subunit protein which is more than twice the size of its E.coli counterpart (EcoL24). Sequence analysis identified two transcript variants that encode the same protein. [provided by Ref- Seq, Jul 2008], Product: Liquid in PBS containing 50% glycerol, 0.5% BSA and 0.02% sodium azide. Immunohistochemistry analysis of paraffin-embedded human breast Molecular Weight: carcinoma tissue, using MRPL24 Antibody. The picture on the right is ~ 32 kDa blocked with the synthesized peptide. Swiss-Prot: Q96A35 Purification&Purity: The antibody was affinity-purified from rabbit antiserum by affinity-chromatography using epitope-specific im- munogen. Applications: Western blot analysis of lysates from HUVEC cells, using MRPL24 An- Western Blot: 1/500 - 1/2000. -

YEASTRACT - Genes Grouped by TF, Ordered by the Percentage of Genes Regulated by TF, Relative to the Total Number of Genes in the List 08/07/14 16:17
YEASTRACT - Genes grouped by TF, ordered by the percentage of genes regulated by TF, relative to the total number of genes in the list 08/07/14 16:17 Home > Group Genes by TF > Result Contact Us - Tutorial - Tutorial Genes grouped by TF, ordered by the percentage of genes regulated by TF, relative to the total number of genes in the list Quick search... Search Documented regulations suported by direct or indirect or undefined evidence. Unknown gene/ORF name(s), 'YFL013W-A'. DISCOVERER Transcription Transcriptional Regulatory % ORF/Genes Regulatory Associations: Factor Network - Search for TFs NTG1 YAL045c HAP3 - Search for Genes ATP1 YBL107c ALG1 - Search for Associations YSA1 CNS1 PDB1 TSC10 RER1 YCL049c PRD1 Group genes: AHC2 GET3 BPL1 - Group by TF YDL144c GCV1 PST2 - Group by GO PST1 SED1 YDR262w YDR266c MSW1 FRQ1 Pattern Matching: TSA2 STP1 PHO8 RAD23 - Search by DNA Motif YEL047c CAN1 YEL074w - Find TF Binding Site(s) PMI40 FMP52 CEM1 - Search Motifs on Motifs SER3 SHC1 YER130c Utilities: PDA1 YER189w AGP3 - ORF List ⇔ Gene List DUG1 CDH1 OCH1 - IUPAC Code Generation YGL114w MRM2 CHO2 - Generate Regulation Matrix PMT6 TRX2 YSC84 CHS7 Yap1p 24.9 % YSP1 FAA3 LYS1 GTT1 Retrieve: YJL045w GZF3 LCB3 - Transcription Factors List HXT9 VPS55 HOM6 BAT2 - Upstream Sequence THI11 RSM22 UBA1 - Flat files RHO4 PAM17 YKR070w YKR077w GTT2 YLL067c About Yeastract: ALT1 HOG1 MAS1 - Contact Us ECM38 ILV5 MRPL4 - Cite YEASTRACT TSA1 PRE8 TAF13 VAN1 - Acknowledgments YTA12 NUP53 SPG5 - Credits MRPL24 NIS1 APP1 CPT1 FMP41 MRPL19 YNL208w CWC25 LYP1 TOF1 -

Integrative Genomic Analysis Predicts Causative Cis-Regulatory Mechanisms of the Breast Cancer-Associated Genetic Variant Rs4415084
Author Manuscript Published OnlineFirst on January 19, 2018; DOI: 10.1158/0008-5472.CAN-17-3486 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. Cancer Research Mathematical Oncology Title: Integrative genomic analysis predicts causative cis-regulatory mechanisms of the breast cancer-associated genetic variant rs4415084 Authors and affiliations: Yi Zhang1,2*, Mohith Manjunath2*, Shilu Zhang3, Deborah Chasman3, Sushmita Roy3,4, and Jun S. Song2,5 *Equal contribution 1Department of Bioengineering, 2Carl R. Woese Institute for Genomic Biology, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA. 3Wisconsin Institute for Discovery, 4Department of Biostatistics and Medical Informatics, University of Wisconsin–Madison, Madison, WI 53792, USA. 5Department of Physics, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA. Running title: Regulatory genomics of GWAS SNPs Abbreviations: GWAS - genome-wide association study; eQTL - expression quantitative trait loci; ASE - allele-specific expression; TF - transcription factor; LD - linkage disequilibrium; FPKM - fragments per kilobase of transcript per million mapped reads; LCASE - local chromosome allele-specific expression; DHS - DNase I hypersensitive sites; PWM - position weight matrix; TCGA - The Cancer Genome Atlas; ER+ - estrogen receptor positive; TAD - topologically associated domain; MAF - minor allele frequency; RPKM - reads per kilobase of transcript per million mapped reads; SNP - single nucleotide polymorphism; MAPQ - mapping quality; ChIP-seq - chromatin immunoprecipitation sequencing; ASB - allele-specific binding; ncRNA - non-coding RNA; TSS - transcription start site; DNase-seq - DNase I 1 Downloaded from cancerres.aacrjournals.org on September 29, 2021. © 2018 American Association for Cancer Research. Author Manuscript Published OnlineFirst on January 19, 2018; DOI: 10.1158/0008-5472.CAN-17-3486 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. -
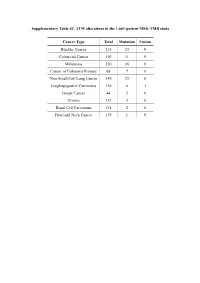
Supplementary Table S1. ATM Alterations in the 1,661-Patient MSK-TMB Study
Supplementary Table S1. ATM alterations in the 1,661-patient MSK-TMB study Cancer Type Total Mutation Fusion Bladder Cancer 215 23 0 Colorectal Cancer 110 11 0 Melanoma 320 26 0 Cancer of Unknown Primary 88 7 0 Non-Small Cell Lung Cancer 350 23 0 Esophagogastric Carcinoma 126 6 1 Breast Cancer 44 2 0 Glioma 117 3 0 Renal Cell Carcinoma 151 2 0 Head and Neck Cancer 139 1 0 Supplementary Table S2. ATM alternations in the 10,945-patient MSK-IMPACT study Cancer Type Total Mutation Deep deletion Amplification Multi-alterations Fusion Small Bowl Cancer 35 7 0 0 0 0 Skin Cancer, Non-Melaoma 148 19 1 0 0 0 Bladder Cancer 423 45 2 0 0 0 Endometrial Cancer 218 24 0 0 0 0 Hepatobiliary Cancer 355 27 0 0 0 0 Colorectal Cancer 1007 75 0 0 0 1 Mature B-Cell Neoplasms 134 8 0 0 2 0 Non-Small Cell Lung Cancer 1668 120 0 2 1 0 Melanoma 365 23 0 0 0 0 Appendiceal Cancer 79 4 0 0 0 0 Small Cell Lung Cancer 82 4 0 0 0 0 Prostate Cancer 717 25 7 1 0 0 Histiocytosis 22 1 0 0 0 0 Salivary Gland Cancer 114 5 0 0 0 0 Thyroid Cancer 231 10 0 0 0 0 Cancer of Unknown Primary 186 8 0 0 0 0 Breast Cancer 1324 48 3 2 0 0 Adrenocortical Carcinoma 25 1 0 0 0 0 Mature T and NK Neoplasms 29 1 0 0 0 0 Renal Cell Carcinoma 361 12 0 0 0 0 Head and Neck Cancer 186 14 0 0 0 1 Pancreatic Cancer 502 69 7 2 0 0 Glioma 553 14 1 0 0 0 Soft Tissue Sarcoma 443 13 1 0 0 0 Esophagogastric Cancer 341 8 1 0 0 0 Peripheral Nervous System 80 0 2 0 0 0 Germ Cell Tumor 288 2 0 1 0 1 Ovarian Can 244 2 1 0 0 0 Uterine Sarcoma 93 1 0 0 0 0 Mesothelioma 107 1 0 0 0 0 Bone Cancer 134 1 0 0 0 0 Gastrointestinal Stromal Ca 137 0 0 0 0 1 Supplementary Table S3. -

Supplementary Dataset S2
mitochondrial translational termination MRPL28 MRPS26 6 MRPS21 PTCD3 MTRF1L 4 MRPL50 MRPS18A MRPS17 2 MRPL20 MRPL52 0 MRPL17 MRPS33 MRPS15 −2 MRPL45 MRPL30 MRPS27 AURKAIP1 MRPL18 MRPL3 MRPS6 MRPS18B MRPL41 MRPS2 MRPL34 GADD45GIP1 ERAL1 MRPL37 MRPS10 MRPL42 MRPL19 MRPS35 MRPL9 MRPL24 MRPS5 MRPL44 MRPS23 MRPS25 ITB ITB ITB ITB ICa ICr ITL original ICr ICa ITL ICa ITL original ICr ITL ICr ICa mitochondrial translational elongation MRPL28 MRPS26 6 MRPS21 PTCD3 MRPS18A 4 MRPS17 MRPL20 2 MRPS15 MRPL45 MRPL52 0 MRPS33 MRPL30 −2 MRPS27 AURKAIP1 MRPS10 MRPL42 MRPL19 MRPL18 MRPL3 MRPS6 MRPL24 MRPS35 MRPL9 MRPS18B MRPL41 MRPS2 MRPL34 MRPS5 MRPL44 MRPS23 MRPS25 MRPL50 MRPL17 GADD45GIP1 ERAL1 MRPL37 ITB ITB ITB ITB ICa ICr original ICr ITL ICa ITL ICa ITL original ICr ITL ICr ICa translational termination MRPL28 MRPS26 6 MRPS21 PTCD3 C12orf65 4 MTRF1L MRPL50 MRPS18A 2 MRPS17 MRPL20 0 MRPL52 MRPL17 MRPS33 −2 MRPS15 MRPL45 MRPL30 MRPS27 AURKAIP1 MRPL18 MRPL3 MRPS6 MRPS18B MRPL41 MRPS2 MRPL34 GADD45GIP1 ERAL1 MRPL37 MRPS10 MRPL42 MRPL19 MRPS35 MRPL9 MRPL24 MRPS5 MRPL44 MRPS23 MRPS25 ITB ITB ITB ITB ICa ICr original ICr ITL ICa ITL ICa ITL original ICr ITL ICr ICa translational elongation DIO2 MRPS18B MRPL41 6 MRPS2 MRPL34 GADD45GIP1 4 ERAL1 MRPL37 2 MRPS10 MRPL42 MRPL19 0 MRPL30 MRPS27 AURKAIP1 −2 MRPL18 MRPL3 MRPS6 MRPS35 MRPL9 EEF2K MRPL50 MRPS5 MRPL44 MRPS23 MRPS25 MRPL24 MRPS33 MRPL52 EIF5A2 MRPL17 SECISBP2 MRPS15 MRPL45 MRPS18A MRPS17 MRPL20 MRPL28 MRPS26 MRPS21 PTCD3 ITB ITB ITB ITB ICa ICr ICr ITL original ITL ICa ICa ITL ICr ICr ICa original -

A Homozygous MRPL24 Mutation Causes a Complex Movement Disorder and Affects the Mitoribosome Assembly T
Neurobiology of Disease 141 (2020) 104880 Contents lists available at ScienceDirect Neurobiology of Disease journal homepage: www.elsevier.com/locate/ynbdi A homozygous MRPL24 mutation causes a complex movement disorder and affects the mitoribosome assembly T Michela Di Nottiaa,1, Maria Marcheseb,1, Daniela Verrignia, Christian Daniel Muttic, Alessandra Torracoa, Romina Olivad, Erika Fernandez-Vizarrac, Federica Moranib, Giulia Trania, Teresa Rizzaa, Daniele Ghezzie,f, Anna Ardissoneg,h, Claudia Nestib, Gessica Vascoi, Massimo Zevianic, Michal Minczukc, Enrico Bertinia, Filippo Maria Santorellib,2, ⁎ Rosalba Carrozzoa, ,2 a Unit of Muscular and Neurodegenerative Disorders, Laboratory of Molecular Medicine, Bambino Gesù Children's Hospital, IRCCS, Rome, Italy b Molecular Medicine & Neurogenetics, IRCCS Fondazione Stella Maris, Pisa, Italy c MRC Mitochondrial Biology Unit, University of Cambridge, Cambridge, UK d Department of Sciences and Technologies, University Parthenope of Naples, Naples, Italy e Unit of Medical Genetics and Neurogenetics, Fondazione IRCCS Istituto Neurologico Carlo Besta, Milan, Italy f Department of Pathophysiology and Transplantation, University of Milan, Milan, Italy g Child Neurology Unit, Fondazione IRCCS Istituto Neurologico Carlo Besta, Milan, Italy h Department of Molecular and Translational Medicine DIMET, University of Milan-Bicocca, Milan, Italy i Department of Neurosciences, IRCCS Bambino Gesù Children Hospital, Rome, Italy ARTICLE INFO ABSTRACT Keywords: Mitochondrial ribosomal protein large 24 (MRPL24) is 1 of the 82 protein components of mitochondrial ribo- Mitochondrial disorders somes, playing an essential role in the mitochondrial translation process. Movement disorder We report here on a baby girl with cerebellar atrophy, choreoathetosis of limbs and face, intellectual dis- MRPL24 ability and a combined defect of complexes I and IV in muscle biopsy, caused by a homozygous missense mu- Mitoribosomes tation identified in MRPL24. -

Microarray Comparative Genomic Hybridization Analysis of 59 Patients with Schizophrenia
J Hum Genet (2008) 53:914–919 DOI 10.1007/s10038-008-0327-6 ORIGINAL ARTICLE Microarray comparative genomic hybridization analysis of 59 patients with schizophrenia Takeshi Mizuguchi Æ Ryota Hashimoto Æ Masanari Itokawa Æ Akira Sano Æ Osamu Shimokawa Æ Yukiko Yoshimura Æ Naoki Harada Æ Noriko Miyake Æ Akira Nishimura Æ Hirotomo Saitsu Æ Nadiya Sosonkina Æ Norio Niikawa Æ Hiroshi Kunugi Æ Naomichi Matsumoto Received: 18 April 2008 / Accepted: 8 July 2008 / Published online: 7 August 2008 Ó The Japan Society of Human Genetics and Springer 2008 Abstract Schizophrenia is a common psychiatric disor- that genome-wide copy number survey should be consid- der with a strong genetic contribution. Disease-associated ered in genetic studies of schizophrenia. chromosomal abnormalities in this condition may provide important clues, such as DISC1. In this study, 59 schizo- Keywords Schizophrenia Á Chromosomal abnormality Á phrenia patients were analyzed by microarray comparative Array comparative genomic hybridization Á genomic hybridization (CGH) using custom bacterial arti- Copy number variation ficial chromosome (BAC) microarray (4,219 BACs with 0.7-Mb resolution). Chromosomal abnormalities were found in six patients (10%): 46,XY,der(13)t(12;13)(p12.1; Introduction p11).ish del(5)(p11p12); 46,XY, ish del(17)(p12p12); 46,XX.ish dup(11)(p13p13); and 46,X,idic(Y)(q11.2); and Schizophrenia is a common psychiatric disorder involving in two cases, mos 45,X/46XX. Autosomal abnormalities in approximately 1% of the population worldwide. Family, three cases are likely to be pathogenic, and sex chromo- twin, and adoption studies suggest genetic factors con- some abnormalities in three follow previous findings.