Identification and Monitoring of Amomi Fructus and Its Adulterants Based
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

11Th Flora Malesina Symposium, Brunei Darussalm, 30 June 5 July 2019 1
11TH FLORA MALESINA SYMPOSIUM, BRUNEI DARUSSALM, 30 JUNE 5 JULY 2019 1 Welcome message The Universiti Brunei Darussalam is honoured to host the 11th International Flora Malesiana Symposium. On behalf of the organizing committee it is my pleasure to welcome you to Brunei Darussalam. The Flora Malesiana Symposium is a fantastic opportunity to engage in discussion and sharing information and experience in the field of taxonomy, ecology and conservation. This is the first time that a Flora Malesiana Symposium is organized in Brunei Darissalam and in the entire island of Borneo. At the center of the Malesian archipelago the island of Borneo magnifies the megadiversity of this region with its richness in plant and animal species. Moreover, the symposium will be an opportunity to inspire and engage the young generation of taxonomists, ecologists and conservationists who are attending it. They will be able to interact with senior researchers and get inspired with new ideas and develop further collaboration. In a phase of Biodiversity crisis, it is pivotal the understanding of plant diversity their ecology in order to have a tangible and successful result in the conservation action. I would like to thank the Vice Chancellor of UBD for supporting the symposium. In the last 6 months the organizing committee has worked very hard for making the symposium possible, to them goes my special thanks. I would like to extend my thanks to all the delegates and the keynote speakers who will make this event a memorable symposium. Dr Daniele Cicuzza Chairperson of the 11th International Flora Malesiana Symposium UBD, Brunei Darussalam 11TH FLORA MALESINA SYMPOSIUM, BRUNEI DARUSSALM, 30 JUNE 5 JULY 2019 2 Organizing Committee Adviser Media and publicity Dr. -

Amomum Foetidum (Zingiberaceae), a New Species from Northeast Thailand
Taiwania 65(3): 364‒370, 2020 DOI: 10.6165/tai.2020.65.364 Amomum foetidum (Zingiberaceae), a new species from Northeast Thailand Thawatphong BOONMA1, Surapon SAENSOUK2,*, Piyaporn SAENSOUK3 1. Brio Garden 53 M.5 Ban Mai Village, Phikun Ok, Ban Na District, Nakhon Nayok, Thailand, 26110. 2. Plant and Invertebrate Taxonomy and Its Applications Unit Group, WalaiRukhavej Botanical Research Institute, Mahasarakham University, Kantarawichai District, Maha Sarakham, Thailand, 44150. 3. Plant and Invertebrate Taxonomy and Its Applications Unit Group, Department of Biology, Faculty of Science, Mahasarakham University, Maha Sarakham, Thailand, 44150. *Corresponding author’s email: [email protected] (Manuscript received 13 April 2020; Accepted 5 July 2020; Online published 13 July 2020) ABSTRACT: Amomum foetidum (Zingiberaceae), a new species from Northeast Thailand is here described, illustrated and photographed. The key to three species of Amomum which cited and treated in this paper is provided. KEY WORDS: Amomum cinnamomeum, Amomum foetidum, new species, Northeast, Thailand, Zingiberaceae. INTRODUCTION previously have been classified in Elettariopsis genus; Amomum curtisii (Baker) Škorničk. & Hlavatá (2018) ≡ The genus Amomum Roxb. is in the subfamily Elettariopsis curtisii Baker (1892) = Elettariopsis Alpinioideae, tribe Alpinieae of the Zingiberaceae serpentina Baker (1892). It did not match with any family (Kress et al., 2002). The genus as currently existing species and treated it as new species, described, circumscribed is distributed from Sri Lanka and India illustrated, and photographed under the name of through SE Asia to New Guinea, the Bismarck Amomum foetidum sp. nov. Archipelago and Australia (Mabberley, 2008). The earliest studies include the phylogenetic analysis by TAXONOMIC TREATMENT Kress et al. -
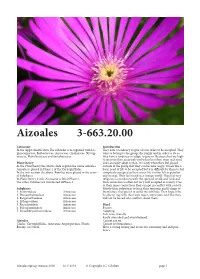
Aizoales 3-663.20.00
Aizoales 3-663.20.00 Taxonomy Introduction In the Apg2 classifcation Te suborder is recognised with Lo- Tey have a tendency to give a lot in order to be accepted. Tey phiocarpaceae, Barbeuiaceae, Aizoaceae, Gisekiaceae, Nyctag- want to belong to the group, the family and in order to do so inaceae, Phytolaccaceae and Sarcobataceae. they have a tendency to adapt, to give in. Because they are high- ly sensitive they accurately feel what the others want and need Plant theory and can easily adapt to that. It is only when they feel placed In the Plant theory the above clade is given the name Aizoales. outside of the group that they can become angry. It feels like a Aizoales is placed in Phase 2 of the Caryophyllidae. basic need of life to be accepted but it is difcult for them to feel In the frst version the above Families were placed in the sever- completely accepted as their inner life is ofen felt as peculiar al Subphases. and strange. Tey feel weird in a strange world. Tey feel very In Plant theory 2 only Aizoaceae is lef inPhase 2. religious, a connection with the spiritual world and God and Te other Families are transferred toPhase 3. that connection is ofen not very well accepted in society. Due to their inner convictions they can get in confict with society. Subphases Mostly their solution is to keep their opinions and feelings to 1. Sesuvioideae Aizoaceae themselves; they prefer to avoid the conficts. Tey hope to be 2. Drosanthemoideae Aizoaceae be able to stay with their own inner convictions and that they 3. -

Setting the Priority Medicinal Plants for Conservation in Indonesia Cahyaningsih, Ria; Magos Brehm, Joana; Maxted, Nigel
University of Birmingham Setting the priority medicinal plants for conservation in Indonesia Cahyaningsih, Ria; Magos Brehm, Joana; Maxted, Nigel DOI: 10.1007/s10722-021-01115-6 License: Creative Commons: Attribution (CC BY) Document Version Publisher's PDF, also known as Version of record Citation for published version (Harvard): Cahyaningsih, R, Magos Brehm, J & Maxted, N 2021, 'Setting the priority medicinal plants for conservation in Indonesia', Genetic Resources and Crop Evolution, vol. 68, no. 5, pp. 2019-2050. https://doi.org/10.1007/s10722-021-01115-6 Link to publication on Research at Birmingham portal General rights Unless a licence is specified above, all rights (including copyright and moral rights) in this document are retained by the authors and/or the copyright holders. The express permission of the copyright holder must be obtained for any use of this material other than for purposes permitted by law. •Users may freely distribute the URL that is used to identify this publication. •Users may download and/or print one copy of the publication from the University of Birmingham research portal for the purpose of private study or non-commercial research. •User may use extracts from the document in line with the concept of ‘fair dealing’ under the Copyright, Designs and Patents Act 1988 (?) •Users may not further distribute the material nor use it for the purposes of commercial gain. Where a licence is displayed above, please note the terms and conditions of the licence govern your use of this document. When citing, please reference the published version. Take down policy While the University of Birmingham exercises care and attention in making items available there are rare occasions when an item has been uploaded in error or has been deemed to be commercially or otherwise sensitive. -

Process Optimization of Microwave Assisted Simultaneous Distillation and Extraction from Siam Cardamom Using Response Surface Methodology
processes Article Process Optimization of Microwave Assisted Simultaneous Distillation and Extraction from Siam cardamom using Response Surface Methodology Panawan Suttiarporn 1 , Nalin Wongkattiya 2 , Kittisak Buaban 2 , Pisit Poolprasert 3 and Keerati Tanruean 4,* 1 Faculty of Science, Energy and Environment, King Mongkut’s University of Technology North Bangkok, Rayong Campus, Rayong 21120, Thailand; [email protected] 2 Program in Biotechnology, Faculty of Science, Maejo University, Chiang Mai 50290, Thailand; [email protected] (N.W.); [email protected] (K.B.) 3 Program in Biology, Faculty of Science and Technology, Pibulsongkram Rajabhat University, Phitsanulok 65000, Thailand; [email protected] 4 Program in Biotechnology, Faculty of Science and Technology, Pibulsongkram Rajabhat University, Phitsanulok 65000, Thailand * Correspondence: [email protected] Received: 20 March 2020; Accepted: 8 April 2020; Published: 11 April 2020 Abstract: The main goal of the research was to optimize microwave-assisted simultaneous distillation and extraction (MA-SDE) using response surface methodology (RSM), based on Box–Behnken design (BBD). A process was designed to extract the essential oil from the leaf sheath of Siam cardamom. The experimental data were fitted to quadratic equations, and the experiment conditions for optimal extraction of 1,8-cineole were extraction time 87.68 min, material-to-water ratio 1:13.18 g/mL and microwave power 217.77 W. Under such conditions, the content of 1,8-cineole was 157.23 4.23 µg/g, which matched with the predicted value. GC–MS results indicated the presence ± of predominant oxygenated monoterpenes including 1,8-cineole (20.63%), iso-carveol (14.30%), cis-p-mentha-1(7),8-dien-2-ol (12.27%) and trans-p-2,8-menthadien-1-ol (9.66%), and oxygenated contents were slightly higher in the MA-SDE and extraction compared to usual SDE. -

Chemotaxonomic Monitoring of Genetically Authenticated Amomi Fructus Using High-Performance Liquid Chromatography–Diode Array Detector with Chemometric Analysis
molecules Article Chemotaxonomic Monitoring of Genetically Authenticated Amomi Fructus Using High-Performance Liquid Chromatography–Diode Array Detector with Chemometric Analysis 1, 2, 3 4 Eui-Jeong Doh y , Guemsan Lee y , Hyun-Jong Jung , Kang-Beom Kwon and Jung-Hoon Kim 5,* 1 Research Center of Traditional Korean Medicine, Wonkwang University, Iksan 54538, Korea; [email protected] 2 Department of Herbology, College of Korean Medicine, Wonkwang University, Iksan 54538, Korea; rasfi[email protected] 3 Department of Diagnostics, College of Korean Medicine, Wonkwang University, Iksan 54538, Korea; [email protected] 4 Department of Korean Medicinal Physiology, College of Korean Medicine, Wonkwang University, Iksan 54538, Korea; [email protected] 5 Division of Pharmacology, School of Korean Medicine, Pusan National University, Yangsan 50612, Korea * Correspondence: [email protected]; Tel.: +82-51-510-8456 These authors contributed equally to this work. y Received: 11 September 2020; Accepted: 6 October 2020; Published: 7 October 2020 Abstract: Amomi Fructus is widely used to treat digestive disorders, and Amomum villosum, A. villosum var. xanthioides, and A. longiligulare are permitted medicinally in national pharmacopeias. However, there are a variety of adulterants present in herbal markets owing to their morphological similarities to the genuine Amomum species. Forty-two Amomi Fructus samples from various origins were identified using internal transcribed spacer and chloroplast barcoding analyses, and then their chromatographic profiles were compared using chemometric analysis for chemotaxonomic monitoring. Among the Amomi Fructus samples, A. villosum, A. longiligulare, A. ghaticum, and A. microcarpum were confirmed as single Amomum species, whereas a mixture of either these Amomum species or with another Amomum species was observed in 15 samples. -

Comparison and Phylogenetic Analyses of Nine Complete Chloroplast Genomes of Zingibereae
Article Comparison and Phylogenetic Analyses of Nine Complete Chloroplast Genomes of Zingibereae Heng Liang 1,2 and Juan Chen 1,* 1 Key Laboratory of Plant Resources Conservation and Sustainable Utilization/Guangdong Provincial Key Laboratory of Digital Botanical Garden, South China Botanical Garden, Chinese Academy of Sciences, Guangzhou 510650, China; [email protected] 2 College of Life Science, Sichuan Agricultural University, Ya’an 625014, China * Correspondence: [email protected] Abstract: Zingibereae is a large tribe in the family Zingiberaceae, which contains plants with impor- tant medicinal, edible, and ornamental values. Although tribes of Zingiberaceae are well circum- scribed, the circumscription of many genera within Zingibereae and the relationships among them remain elusive, especially for the genera of Boesenbergia, Curcuma, Kaempferia and Pyrgophyllum. In this study, we investigated the plastome variation in nine species representing five genera of Zingibereae. All plastomes showed a typical quadripartite structure with lengths ranging from 162,042 bp to 163,539 bp and contained 132–134 genes, consisting of 86–88 coding genes, 38 transfer RNA genes and eight ribosomal RNA genes. Moreover, the characteristics of the long repeats sequences and simple sequence repeats (SSRs) were detected. In addition, we conducted phylogenomic analyses of the Zingibereae and related taxa with plastomes data from additional 32 species from Genbank. Our results confirmed that Stahlianthus is closely related to Curcuma, supporting the idea of merging it into Curcuma. Kaempferia, Boesenbergia and Zingiber were confirmed as close relatives and grouped together as the Kaempferia group. Pyrgophyllum is not allied with the Curcuma clade but instead is Citation: Liang, H.; Chen, J. -

Thai Zingiberaceae of Pharmacological Interest
Supplementary Materials Thai Zingiberaceae of pharmacological interest Phumthum, M. and Balslev, H. Table S1 Ethnomedicinal uses of gingers in Thailand. Plant Name Uses (Preparation/Application) References Alpinia Alpinia calcarata (Haw.) Roscoe Diarrhea (dc/oi), diuretic agents (dc/oi) Anderson (1993), Srithi (2012) Alpinia conchigera Griff. Asthma (cr/al), diabetes mellitus (un/un) Chuakul et al. (2004), Chuakul and Boonpleng (2003) Alpinia galanga (L.) Willd. Amenorrhoea (un/un), anaesthesia Chuakul (2012), Chuakul and (sw/ba), anthelminthic (un/un), back Boonpleng (2003), Chuakul et al. and waist pain (dc/oi), carminative (2002a), Gunsuwan (2011), Inta (2008), (un/un), colitis (cf/oi), congestion Jitjum et al. (2019), Kaewwongsiri and (un/un), convulsion (un/un), cough Saiyapan (2004), Khonkayan et al. (dc/oi), dermatophytosis (un/un), (2019), Khuankaew (2014), Neamsuvan diarrhea (np/oi), dizziness (cr/sm), fever (2013a), Nuammee (2012), Pannet (dc/oi), flatulence (dc/ba), headache (2008), Panyadee et al. (2019), Ponpim (dc/oi), hypertension (un/un), insect (1996), Pantarod (2002), Purintavaragul bites and stings (dc/oi), laxative (un/un) et al. (2012), Sonsupub (2010), Srithi muscular relaxation (un/un), paralysis (2012), Thaenkam et al. (2019), (un/un), peptic ulcers (un/un), Winijchaiyanan (1995) postpartum abdominal pain (un/un), postpartum nervous (dc/ba), postpartum tonic (un/un), toothache (dc/oi), venomous animal bites (cr/al) Alpinia malaccensis (Burm.f.) Abdominal pain (np/oi), cough (dc/oi), Anderson (1993), Kaewsangsai (2017), Roscoe dermatophytosis (po/un), flatulence Kantasrila (2016), Neamsuvan et al. (np/oi), food poisoning (np/oi), (2014), Neamsuvan and Tuntien (2015), indigestion (dc/oi), pruritus (dc/oi), Panyadee et al. (2019), Srisanga et al. -
Taxonomic Studies on Amomum Roxburgh S.L. (Zingiberaceae) in Myanmarⅰ: Two New Species and Two New Records for the Flora of Myanmar
Phytotaxa 418 (2): 158–170 ISSN 1179-3155 (print edition) https://www.mapress.com/j/pt/ PHYTOTAXA Copyright © 2019 Magnolia Press Article ISSN 1179-3163 (online edition) https://doi.org/10.11646/phytotaxa.418.2.2 Taxonomic studies on Amomum Roxburgh s.l. (Zingiberaceae) in MyanmarⅠ: Two new species and two new records for the flora of Myanmar HONG-BO DING1,2, SHI-SHUN ZHOU1,2, BIN YANG1,2, REN LI1,2, MYA BHONE MAW1,2, KYAW WIN MAUNG3 & YUN-HONG TAN1,2* 1Southeast Asia Biodiversity Research Institute, Chinese Academy of Sciences & Center for Integrative Conservation, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Menglun, Mengla, Yunnan 666303, P.R. China 2Center of Conservation Biology, Core Botanical Gardens, Chinese Academy of Sciences, Menglun, Mengla,Yunnan 666303, P.R. China 3Forest Research Institute, Forest Department, Ministry of Environmental Conservation and Forestry, Yezin, Nay Pyi Taw 05282, Myanmar *Author for correspondence. E-mail: [email protected] Abstract Two taxa of Amomum (Zingiberaceae), Amomum erythranthum and Amomum ampliflorum, from Putao, Kachin State of Northern Myanmar are described and illustrated as new to science. Amomum erythranthum is morphologically similar to A. subulatum and A. nimkeyense in having similar yellow flowers, but can be distinguished by its reddish floral tube, red anther connective, red and pubescent fruit. Amomum ampliflorum is similar to A. maximum, A. dealbatum and A. odontocarpum in white flower, but differs in its longer inflorescence and much larger flower. Two species, Amomum pauciflorum and Wurfbainia microcarpum are recorded for the flora of Myanmar for the first time. Data on ecology, phenology, distribution, conservation status, similarities to the related taxa, as well as colour photographs and line drawings of the type, and voucher specimens are provided for all reported taxa. -
Zingiberaceae) in Myanmar II
A peer-reviewed open-access journal PhytoKeys 138:Taxonomic 139–153 (2020) studies on Amomum Roxburgh s.l. (Zingiberaceae) in Myanmar II... 139 doi: 10.3897/phytokeys.138.38736 RESEARCH ARTICLE http://phytokeys.pensoft.net Launched to accelerate biodiversity research Taxonomic studies on Amomum Roxburgh s.l. (Zingiberaceae) in Myanmar II: one new species and five new records for the flora of Myanmar Hong-Bo Ding1,2, Bin Yang1,2, Mya Bhone Maw1,2, Pyae Pyae Win3, Yun-Hong Tan1,2 1 Southeast Asia Biodiversity Research Institute, Chinese Academy of Sciences & Center for Integrative Conser- vation, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Menglun, Mengla, Yunnan 666303, China 2 Center of Conservation Biology, Core Botanical Gardens, Chinese Academy of Sciences, Menglun, Mengla,Yunnan 666303, China 3 Forest Research Institute, Forest Department, Ministry of Envi- ronmental Conservation and Forestry, Yezin, Nay Pyi Taw 05282, Myanmar Corresponding author: Yun-Hong Tan ([email protected]) Academic editor: Xiao-Hua Jin | Received 3 August 2019 | Accepted 26 November 2019 | Published 10 January 2020 Citation: Ding H-B, Yang B, Maw MB, Win PP, Tan Y-H (2020) Taxonomic studies on Amomum Roxburgh s.l. (Zingiberaceae) in Myanmar II: one new species and five new records for the flora of Myanmar. In: Jin X-H, Xia N-H, Tan Y-H (Eds) Plant diversity of Southeast Asia-II. PhytoKeys 138: 139–153. https://doi.org/10.3897/phytokeys.138.38736 Abstract In the course of a study of Amomum s.l. (Zingiberaceae) in Myanmar, Amomum schistocalyx Y.H. Tan & H.B. Ding, from Htamanti Wildlife Sanctuary, Sangaing Region of Northern Myanmar is described and illustrated as new to science here. -

Species Richness and Endemism of Zingiberaceae in Cinchona Forest Reserve, Lantapan, Bukidnon, Philippines
JOURNAL OF TROPICAL LIFE SCIENCE 2020, Vol. 10, No. 2, 175 – 180 http://dx.doi.org/10.11594/jtls.10.02.10 Research Article Species Richness and Endemism of Zingiberaceae in Cinchona Forest Reserve, Lantapan, Bukidnon, Philippines John Austin Lennox Faro Jayme 1, Noe Polo Mendez 2*, Rainear Auxtero Mendez 2, Daniel F. Somera 3, Alma B. Mohagan 2, 4 1 Senior High School, Central Mindanao University, University Town, Musuan 8710 Bukidnon, Philippines 2 Center for Biodiversity Research and Extension in Mindanao (CEBREM), Central Mindanao University, University Town, Musuan 8710 Bukidnon, Philippines 3 Protected Area Superintendent - Mt. Kitanglad Range Natural Park (MKRNP), Malaybalay City, Bukidnon, Philippines 4 Department of Biology, College of Arts and Sciences, Central Mindanao University, University Town, Musuan 8710 Bukidnon, Philippines Article history: ABSTRACT Submission October 2019 Revised November 2019 This study was carried out to provide information on species richness and Accepted April 2020 endemism of Zingiberaceae in Cinchona Forest Reserve, Kaatuan, Lanta- pan, Bukidnon, Philippines. Transect walks, opportunistic sampling and *Corresponding author: collection within the sampling quadrats were conducted along established E-mail: [email protected] forest trails to collect ginger species. A total of 11 species of Zingibera- ceae were documented belonging to two subfamilies (Alpinioideae and Zingiberoideae) and three tribes (Alpinieae, Hedychieae, and Zingi- bereae). The species recorded include Adelmeria alpina Elmer, Alpinia haenkei C.Presl, A. rufa C.Presl, Etlingera fimbriobracteata (K.Schum.) R.M.Sm., E. pubimarginata (Elmer) A.D.Poulsen, Hedychium philip- pinense K.Schum., Hornstedtia conoidea Ridl., H. lophophora Ridl., Meistera muricarpa (Elmer) Škorničk. & M.F.Newman, Zingiber banahaoense Mood & Theilade, and Zingiber sp. -

Plants of Cát Tiên National Park Danh Lục Thực Vật Vườn Quốc Gia Cát Tiên
Plants of Cát Tiên National Park 04 August 2021 Danh lục thực vật Vườn Quốc Gia Cát Tiên Higher Bộ Family Chi - Loài Authority Ngành / Lớp / Phân họ Họ Rec. No.* Clas. Order Species (& Sub-family) ssp., var., syn. etc. & notes TÊN VIỆT NAM Ds Cd Mã số Clade: Embryophyta Nhánh: Thực vật có phôi (Division) Marchantiophyta Liverworts Ngành Rêu tản (Division) Anthocerotophyta Hornworts Ngành Rêu sừng (Division) Bryophyta Mosses Ngành Rêu Tracheophyta (Vascular plants) Thực vật có mạch (Division) Lycopodiophyta clubmosses, etc Ngành Thạch tùng Lycopodiales Huperziaceae firmosses Họ Thạch sam Huperzia carinata (Poir.) Trevis Phlegmariurus is the tropical sub-genus Thạch tùng sóng K C - T 4 Huperzia squarrosa (Forst.) Trevis Thạch tùng vảy K T 12 Huperzia obvalifolia (Bon.) Thạch tùng xoan ngược K C - T 8 Huperzia phlegmaria (L.) Roth Râu cây K C - T 9 Lycopodiaceae clubmosses Họ Thạch tùng Lycopodiella cernua (L.) Franco & Vasc Thạch tùng nghiên K T 16 Lycopodiella sp. Thạch tùng K T Selaginellales Selaginellaceae spikemosses Họ quyển bá Selaginella delicatula (Desv) Alst. Quyển bá yếu K T 41 Selaginella rolandi-principis Alston. Hoa đá K T 27 Selaginella willdenowii (Desv.) Baker. Quyển bá Willdenov K T 33 Selaginella chrysorrhizos Spring Quyển bá vàng K 39 Selaginella minutifolia Spring Quyển bá vi diệp K 49 (Division) Pteridophyta (Polypodiophyta) Leptosporangiate ferns Ngành Dương xỉ Class: Marattiopsida Lớp Dương xỉ tòa sen Marattiales Marattiaceae (previously Angiopteridaceae) Họ Dương xỉ tòa sen Angiopteris repandulade Vriese. Ráng hiền dực, Dương xỉ móng trâu K 82 Class: Pteridopsida or Polypodiopsida Lớp Dương xỉ Polypodiales polypod ferns Bộ Dương xỉ Aspleniaceae Họ Can xỉ (tổ diều) Asplenium nidus L.