Chemotaxonomic Monitoring of Genetically Authenticated Amomi Fructus Using High-Performance Liquid Chromatography–Diode Array Detector with Chemometric Analysis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Identification and Monitoring of Amomi Fructus and Its Adulterants Based
molecules Article Identification and Monitoring of Amomi Fructus and its Adulterants Based on DNA Barcoding Analysis and Designed DNA Markers Eui Jeong Doh 1,2 , Jung-Hoon Kim 3 and Guemsan Lee 1,2,* 1 Department of Herbology, College of Korean Medicine, Wonkwang University, Iksan 54538, Korea; [email protected] 2 Research Center of Traditional Korean Medicine, Wonkwang University, Iksan 54538, Korea 3 Division of Pharmacology, School of Korean Medicine, Pusan National University, Yangsan 50612, Korea; [email protected] * Correspondence: rasfi[email protected]; Tel.: +82-63-850-6985 Received: 18 October 2019; Accepted: 13 November 2019; Published: 19 November 2019 Abstract: Amomi Fructus is one of the traditional medicines derived from the ripe fruits of the Zingiberaceae family of plants, which include Amomum villosum, A. villosum var. xanthioides, and A. longiligulare. Owing to their highly similar morphological traits, several kinds of adulterants of Amomi Fructus have been reported. Therefore, accurate and reliable methods of identification are necessary in order to ensure drug safety and quality. We performed DNA barcoding using five regions (ITS, matK, rbcL, rpoB, and trnL-F intergenic spacer) of 23 Amomi Fructus samples and 22 adulterants. We designed specific DNA markers for Amomi Fructus based on the single nucleotide polymorphisms (SNPs) in the ITS. Amomi Fructus was well separated from the adulterants and was classified with the species of origin based on the detected SNPs from the DNA barcoding results. The AVF1/ISR DNA marker for A. villosum produced a 270 bases amplified product, while the ALF1/ISF DNA marker produced a 350 bases product specific for A. -

Download Download
Firenze University Press Caryologia www.fupress.com/caryologia International Journal of Cytology, Cytosystematics and Cytogenetics Karyological studies in thirteen species of Zingiberacaeae from Tripura, North East India Citation: K. Saha, R.K. Sinha, S. Sin- ha (2020) Karyological studies in thir- teen species of Zingiberacaeae from Tripura, North East India. Caryologia Kishan Saha*, Rabindra Kumar Sinha, Sangram Sinha 73(1): 163-178. doi: 10.13128/caryolo- gia-186 Cytogenetics and Plant Biotechnology Laboratory, Department of Botany, Tripura Univer- sity, Suryamaninagar-799022, Tripura, India Received: March 11, 2019 *Corresponding author. E-mail: [email protected] Accepted: February 23, 2020 Abstract. Tripura being a state in North East India belongs to Indo-Burma bio-diver- Published: May 8, 2020 sity hotspot and is considered as a centre of origin of many species of Zingiberaceae. Copyright: © 2020 K. Saha, R. Alpinia calcarata, Alpinia malaccensis, Alpinia nigra, Amomum aromaticum, Amomum Kumar Sinha, S. Sinha. This is an koenigii, Amomum maximum, Curcuma amada, Curcuma caesia, Curcuma longa, Cur- open access, peer-reviewed article cuma picta, Hedychium coccineum, Hedychium coronarium and Hedychium thyrsiforme published by Firenze University Press are found in wild state in different geographical locations of Tripura. Their karyotypes (http://www.fupress.com/caryologia) were analyzed both at interspecific and intraspecific levels. The somatic chromosome and distributed under the terms of the number of Alpinia spp. and Amomum spp. was found to be 2n = 4X = 48. The Cur- Creative Commons Attribution License, cuma spp. represented by C. amada had 2n = 42 chromosomes and three other species which permits unrestricted use, distri- viz., C. -

11Th Flora Malesina Symposium, Brunei Darussalm, 30 June 5 July 2019 1
11TH FLORA MALESINA SYMPOSIUM, BRUNEI DARUSSALM, 30 JUNE 5 JULY 2019 1 Welcome message The Universiti Brunei Darussalam is honoured to host the 11th International Flora Malesiana Symposium. On behalf of the organizing committee it is my pleasure to welcome you to Brunei Darussalam. The Flora Malesiana Symposium is a fantastic opportunity to engage in discussion and sharing information and experience in the field of taxonomy, ecology and conservation. This is the first time that a Flora Malesiana Symposium is organized in Brunei Darissalam and in the entire island of Borneo. At the center of the Malesian archipelago the island of Borneo magnifies the megadiversity of this region with its richness in plant and animal species. Moreover, the symposium will be an opportunity to inspire and engage the young generation of taxonomists, ecologists and conservationists who are attending it. They will be able to interact with senior researchers and get inspired with new ideas and develop further collaboration. In a phase of Biodiversity crisis, it is pivotal the understanding of plant diversity their ecology in order to have a tangible and successful result in the conservation action. I would like to thank the Vice Chancellor of UBD for supporting the symposium. In the last 6 months the organizing committee has worked very hard for making the symposium possible, to them goes my special thanks. I would like to extend my thanks to all the delegates and the keynote speakers who will make this event a memorable symposium. Dr Daniele Cicuzza Chairperson of the 11th International Flora Malesiana Symposium UBD, Brunei Darussalam 11TH FLORA MALESINA SYMPOSIUM, BRUNEI DARUSSALM, 30 JUNE 5 JULY 2019 2 Organizing Committee Adviser Media and publicity Dr. -

Notes on Some Zingibers from West Bengal and Meghalaya, India
Pleione 10(2): 310 - 322. 2016. ISSN: 0973-9467 © East Himalayan Society for Spermatophyte Taxonomy Notes on some Zingibers from West Bengal and Meghalaya, India Sreetama Bhadra and Maumita Bandyopadhyay1 Plant Molecular Cytogenetics Laboratory, Centre of Advanced Study, Department of Botany, University of Calcutta, 35 Ballygunge Circular Road, Kolkata – 700019, West Bengal, India 1 Corresponding author; E-mail: [email protected] [Received 20.08.2016; Revised 12.10.2016; Accepted 29.11.2016; Published 31.12.2016] Abstract West Bengal and Meghalaya encompasses a bulk of Eastern Himalaya and the present study reports the diversity of Zingiberaceae in these two states. A total of 36 species belonging to 12 genera were recorded and morphologically described in the present investigation including their chromosome counts. Key words: Zingiberaceae, West Bengal, Meghalaya, Morphology, Chromosome numbers INTRODUCTION Zingiberaceae, one of the largest monocot families, possess the maximum number of species among all the 8 families of Zingiberales. The highest concentration of zingibers is found in the tropical and sub–tropical regions of the world. These plants generally grow in tropical rain and monsoon forests, in damp and humid conditions (Chen et al. 1989; Jatoi et al. 2007). The centre of diversity of Zingiberaceae is considered to be primarily in South and South–East Asia (Larsen et al. 1998), including the Indo-Malayan region (Chakravorti 1948; Joseph 1998; Larsen et al. 1998; Kress et al. 2002; Islam 2004; Sabu 2006) with a few genera being distributed in the neotropics (South and Central America) and Africa (Smith 1998; Harris et al. 2000). In India, these plants are confined to the Himalayas, specially the North–Eastern parts, and also in the Western Ghats (Joseph 1998; Sabu 2006). -

Ethnomedicine and Materia Medica Used by Kui Traditional Healers in Northeast Thailand
ETHNOMEDICINE AND MATERIA MEDICA USED BY KUI TRADITIONAL HEALERS IN NORTHEAST THAILAND MISS ARIKA VIRAPONGSE A THESIS FOR THE DEGREE OF MASTER OF SCIENCE KHON KAEN UNIVERSITY 2006 ISBN 974-626-847-3 ETHNOMEDICINE AND MATERIA MEDICA USED BY KUI TRADITIONAL HEALERS IN NORTHEAST THAILAND MISS ARIKA VIRAPONGSE A THESIS SUBMITTED IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF MASTER OF SCIENCE IN PHARMACEUTICAL CHEMISTRY AND NATURAL PRODUCTS GRADUATE SCHOOL KHON KAEN UNIVERSITY 2006 ISBN 974-626-847-3 อารีกา วีระพงส. 2549. การแพทยพื้นบานและยาสมุนไพรที่ใชโดยหมอพื้นบานชนชาติกุยใน ภาคตะวันออกเฉียงเหนือของประเทศไทย. วิทยานิพนธปริญญาวิทยาศาสตรมหาบณฑั ิต สาขาวิชาเภสชเคมั ีและผลิตภัณฑธรรมชาติ บัณฑิตวิทยาลัย มหาวิทยาลัยขอนแกน. [ISBN 974-626-847-3] อาจารยที่ปรึกษาวิทยานิพนธ: รศ.ดร. ชยันต พิเชียรสุนทร, ผศ.ดร.จุลรัตน คนศิลป, อ. ประธาน ฦาชา บทคัดยอ ชนชาติกุยเปนกลุมชาติพันธุหนึ่งในหาสิบกลุมชาติพันธุที่ไมใชไตในประเทศไทย และอาจเปน กลุมชาติพันธุแรก ๆ ที่เขามาตั้งถิ่นฐานอาศัยอยูทางตอนใตของภาคตะวันออกเฉียงเหนือของประเทศไทย การศึกษานี้มีวัตถุประสงคเพื่อศึกษาประมวลแนวคิดในเชิงทฤษฎีในระบบการแพทยพื้นบานของกลุมชาติ พันธุกุย และยาสมุนไพรที่ใชบําบัดโรคหรือความเจ็บปวยตาง ๆ กลุมประชากรที่ใชในการศึกษาครั้งนี้เปน หมอพื้นบานชาวกุยจํานวน 388 คน ซึ่งตั้งถิ่นฐานอยูในพื้นที่จังหวัดสุรินทร ศรีสะเกษ และอุบลราชธานี โดยการศึกษาเชิงลึกกับหมอพื้นบานจํานวน 30 คน วิธีวิจัยที่ใชในการเก็บรวบรวมขอมูลประกอบดวยการ ใชความจําแบบเสรี บันทึกขอมูลดานสุขภาพ การสังเกตการณโดยนักวิจัย การสังเกตการณโดยผูเขารวม โครงการวิจัย การเก็บรวบรวมตัวอยางสมุนไพร และการสัมภาษณ ผลที่ไดไดวิเคราะหและประมวลเปน -

Amomum Foetidum (Zingiberaceae), a New Species from Northeast Thailand
Taiwania 65(3): 364‒370, 2020 DOI: 10.6165/tai.2020.65.364 Amomum foetidum (Zingiberaceae), a new species from Northeast Thailand Thawatphong BOONMA1, Surapon SAENSOUK2,*, Piyaporn SAENSOUK3 1. Brio Garden 53 M.5 Ban Mai Village, Phikun Ok, Ban Na District, Nakhon Nayok, Thailand, 26110. 2. Plant and Invertebrate Taxonomy and Its Applications Unit Group, WalaiRukhavej Botanical Research Institute, Mahasarakham University, Kantarawichai District, Maha Sarakham, Thailand, 44150. 3. Plant and Invertebrate Taxonomy and Its Applications Unit Group, Department of Biology, Faculty of Science, Mahasarakham University, Maha Sarakham, Thailand, 44150. *Corresponding author’s email: [email protected] (Manuscript received 13 April 2020; Accepted 5 July 2020; Online published 13 July 2020) ABSTRACT: Amomum foetidum (Zingiberaceae), a new species from Northeast Thailand is here described, illustrated and photographed. The key to three species of Amomum which cited and treated in this paper is provided. KEY WORDS: Amomum cinnamomeum, Amomum foetidum, new species, Northeast, Thailand, Zingiberaceae. INTRODUCTION previously have been classified in Elettariopsis genus; Amomum curtisii (Baker) Škorničk. & Hlavatá (2018) ≡ The genus Amomum Roxb. is in the subfamily Elettariopsis curtisii Baker (1892) = Elettariopsis Alpinioideae, tribe Alpinieae of the Zingiberaceae serpentina Baker (1892). It did not match with any family (Kress et al., 2002). The genus as currently existing species and treated it as new species, described, circumscribed is distributed from Sri Lanka and India illustrated, and photographed under the name of through SE Asia to New Guinea, the Bismarck Amomum foetidum sp. nov. Archipelago and Australia (Mabberley, 2008). The earliest studies include the phylogenetic analysis by TAXONOMIC TREATMENT Kress et al. -
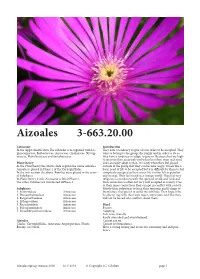
Aizoales 3-663.20.00
Aizoales 3-663.20.00 Taxonomy Introduction In the Apg2 classifcation Te suborder is recognised with Lo- Tey have a tendency to give a lot in order to be accepted. Tey phiocarpaceae, Barbeuiaceae, Aizoaceae, Gisekiaceae, Nyctag- want to belong to the group, the family and in order to do so inaceae, Phytolaccaceae and Sarcobataceae. they have a tendency to adapt, to give in. Because they are high- ly sensitive they accurately feel what the others want and need Plant theory and can easily adapt to that. It is only when they feel placed In the Plant theory the above clade is given the name Aizoales. outside of the group that they can become angry. It feels like a Aizoales is placed in Phase 2 of the Caryophyllidae. basic need of life to be accepted but it is difcult for them to feel In the frst version the above Families were placed in the sever- completely accepted as their inner life is ofen felt as peculiar al Subphases. and strange. Tey feel weird in a strange world. Tey feel very In Plant theory 2 only Aizoaceae is lef inPhase 2. religious, a connection with the spiritual world and God and Te other Families are transferred toPhase 3. that connection is ofen not very well accepted in society. Due to their inner convictions they can get in confict with society. Subphases Mostly their solution is to keep their opinions and feelings to 1. Sesuvioideae Aizoaceae themselves; they prefer to avoid the conficts. Tey hope to be 2. Drosanthemoideae Aizoaceae be able to stay with their own inner convictions and that they 3. -

Setting the Priority Medicinal Plants for Conservation in Indonesia Cahyaningsih, Ria; Magos Brehm, Joana; Maxted, Nigel
University of Birmingham Setting the priority medicinal plants for conservation in Indonesia Cahyaningsih, Ria; Magos Brehm, Joana; Maxted, Nigel DOI: 10.1007/s10722-021-01115-6 License: Creative Commons: Attribution (CC BY) Document Version Publisher's PDF, also known as Version of record Citation for published version (Harvard): Cahyaningsih, R, Magos Brehm, J & Maxted, N 2021, 'Setting the priority medicinal plants for conservation in Indonesia', Genetic Resources and Crop Evolution, vol. 68, no. 5, pp. 2019-2050. https://doi.org/10.1007/s10722-021-01115-6 Link to publication on Research at Birmingham portal General rights Unless a licence is specified above, all rights (including copyright and moral rights) in this document are retained by the authors and/or the copyright holders. The express permission of the copyright holder must be obtained for any use of this material other than for purposes permitted by law. •Users may freely distribute the URL that is used to identify this publication. •Users may download and/or print one copy of the publication from the University of Birmingham research portal for the purpose of private study or non-commercial research. •User may use extracts from the document in line with the concept of ‘fair dealing’ under the Copyright, Designs and Patents Act 1988 (?) •Users may not further distribute the material nor use it for the purposes of commercial gain. Where a licence is displayed above, please note the terms and conditions of the licence govern your use of this document. When citing, please reference the published version. Take down policy While the University of Birmingham exercises care and attention in making items available there are rare occasions when an item has been uploaded in error or has been deemed to be commercially or otherwise sensitive. -

Makalah Jahe
KANDUNGAN OBAT YANG TERDAPAT PADA TUMBUHAN JAHE (ZINGIBERACEAE) Diajukan untuk Memenuhi Salah Satu Tugas Mata Kuliah Morsistum Disusun oleh : YOSSI FEBRIANI NIM 31110052 Farmasi 1A PROGRAM STUDI S1 FARMASI STIKes BHAKTI TUNAS HUSADA TASIKMALAYA 2011 ABSTRAK Zingiberaceae (jahe-jahean) merupakan salah satu kelompok tumbuhan yang telah banyak dimanfaatkan. Kegunaannya tidak hanya sebagai tanaman obat, juga merupakan sumber panghasil minyak esensial dan bahan bumbu. Selain yang sudah dibudidayakan, di hutan-hutan Sumatera Barat masih ditemukan jenis liar Zingiberaceae. Dari studi keanekaragaman yang telah dilakukan, secara keseluruhan ditemukan 17 jenis Zingiberaceae di kawasan Cagar alam Rimbo Panti, Sumatera Barat yakni: Alpinia malaccensis, Amomum gracile, Amomum lappaceum, Amomum testaceum, Amomum sp., Boesenbergia sp., Etlingera megalocheilos, Etlingera sp., Geocharis rubra, Globba aurantiaca, Globba flavibracteata, Globba paniculata, Hedychium longicornutum, Hornstedtia conica, Hornstedtia rubra, Hornstedtia tomentosa dan Zingiber sp. Geocharis rubra merupakan temuan distribusi baru (”new record”) untuk Sumatra. Sedangkan Etlingera sp. diperkirakan jenis baru (”new species”), namun masih diperlukan kajian yang lebih rinci untuk menjelaskan status taksonominya. Kata kunci : Zingiberacae liar, keanekaragaman, Sumatra, “new record”, “new species”. KATA PENGANTAR Puji dan syukur penulis panjatkan kepada Allah SWT yang telah memberikan rahmat-Nya sehingga penulis dapat menyelesaikan laporan penelitian ini tepat pada waktunya. Adapun maksud dari penyusunan laporan ini adalah untuk memenuhi salah satu tugas mata kuliah Morsistum STIKEs Bhakti Tunas Husada. Penyusun menyadari bahwa laporan ini masih jauh dari sempurna. Untuk itu, penyusun sangat mengharapkan saran dan keritik yang sifatnya membangun untuk perbaikan di masa yang akan datang. Penyusun juga menyadari bahwa dalam menyusun laporan ini tidak terlepas dari peran serta dan bantuan dari berbagai pihak, baik bantuan berupa moril maupun materil. -

Process Optimization of Microwave Assisted Simultaneous Distillation and Extraction from Siam Cardamom Using Response Surface Methodology
processes Article Process Optimization of Microwave Assisted Simultaneous Distillation and Extraction from Siam cardamom using Response Surface Methodology Panawan Suttiarporn 1 , Nalin Wongkattiya 2 , Kittisak Buaban 2 , Pisit Poolprasert 3 and Keerati Tanruean 4,* 1 Faculty of Science, Energy and Environment, King Mongkut’s University of Technology North Bangkok, Rayong Campus, Rayong 21120, Thailand; [email protected] 2 Program in Biotechnology, Faculty of Science, Maejo University, Chiang Mai 50290, Thailand; [email protected] (N.W.); [email protected] (K.B.) 3 Program in Biology, Faculty of Science and Technology, Pibulsongkram Rajabhat University, Phitsanulok 65000, Thailand; [email protected] 4 Program in Biotechnology, Faculty of Science and Technology, Pibulsongkram Rajabhat University, Phitsanulok 65000, Thailand * Correspondence: [email protected] Received: 20 March 2020; Accepted: 8 April 2020; Published: 11 April 2020 Abstract: The main goal of the research was to optimize microwave-assisted simultaneous distillation and extraction (MA-SDE) using response surface methodology (RSM), based on Box–Behnken design (BBD). A process was designed to extract the essential oil from the leaf sheath of Siam cardamom. The experimental data were fitted to quadratic equations, and the experiment conditions for optimal extraction of 1,8-cineole were extraction time 87.68 min, material-to-water ratio 1:13.18 g/mL and microwave power 217.77 W. Under such conditions, the content of 1,8-cineole was 157.23 4.23 µg/g, which matched with the predicted value. GC–MS results indicated the presence ± of predominant oxygenated monoterpenes including 1,8-cineole (20.63%), iso-carveol (14.30%), cis-p-mentha-1(7),8-dien-2-ol (12.27%) and trans-p-2,8-menthadien-1-ol (9.66%), and oxygenated contents were slightly higher in the MA-SDE and extraction compared to usual SDE. -

The Taxonomic Investigation on Genus Alpinia Roxb. (Zingiberaceae) from the Siang Valley, Arunachal Pradesh
Bulletin of Arunachal Forest Research, Vol. 33(1), 44-51: 2018 ISSN 0970-9487 THE TAXONOMIC INVESTIGATION ON GENUS ALPINIA ROXB. (ZINGIBERACEAE) FROM THE SIANG VALLEY, ARUNACHAL PRADESH Tatum Mibang*1, Arup K. Das2 1Department of Botany, J.N. College, Pasighat- 791103, Arunachal Pradesh, India 2Department of Botany, Rajiv Gandhi University, Itanagar, Arunachal Pradesh, India E-mail: [email protected] Abstract The Genus Alpinia Roxb. distributes in the tropical and sub-tropical regions of the world. The Siang Valley of Arunachal Pradesh is favorable for the growth of the Genus Alpinia. The extensive taxonomic investigation on the Genus Alpinia in the Siang Valley resulting to 5 (five) species. For better description and understanding, a color photograph of each species are given. Keywords: Alpinia; Taxonomic investigation; Siang Valley INTRODUCTION forest crisscrossed with rivers and streams which is favorable for growth of Zingiberaceae The Genus Alpinia Roxb. consists of 241 species in the Valley. The geographical species (The Plant List, 2010) in World, parameters like altitudes ranges from 130 - mainly distributed in the tropical and sub- 900 m (Ruksin - Pekii Modi), temperature tropical region of South Asian countries. The range from 16 - 34 °C, relative humidity 54 - Genus is characterized by the tall leafy shoot, 98 % and sandy loamy soil enhance the distichous leaves, the plane of distichous is luxurious growth of various Zingiberaceae transverse to the rhizome, terminal species in the Valley. Out of 10 Genera inflorescence with large spectabilis labellum reported (Tatum, 2016) of the family and the staminode absent or rudiment tooth Zingiberaceae from the Siang Valley, Alpinia like or well elaborated horn like arching over is one of the interested genera in the family the basal part of labellum. -

Comparison and Phylogenetic Analyses of Nine Complete Chloroplast Genomes of Zingibereae
Article Comparison and Phylogenetic Analyses of Nine Complete Chloroplast Genomes of Zingibereae Heng Liang 1,2 and Juan Chen 1,* 1 Key Laboratory of Plant Resources Conservation and Sustainable Utilization/Guangdong Provincial Key Laboratory of Digital Botanical Garden, South China Botanical Garden, Chinese Academy of Sciences, Guangzhou 510650, China; [email protected] 2 College of Life Science, Sichuan Agricultural University, Ya’an 625014, China * Correspondence: [email protected] Abstract: Zingibereae is a large tribe in the family Zingiberaceae, which contains plants with impor- tant medicinal, edible, and ornamental values. Although tribes of Zingiberaceae are well circum- scribed, the circumscription of many genera within Zingibereae and the relationships among them remain elusive, especially for the genera of Boesenbergia, Curcuma, Kaempferia and Pyrgophyllum. In this study, we investigated the plastome variation in nine species representing five genera of Zingibereae. All plastomes showed a typical quadripartite structure with lengths ranging from 162,042 bp to 163,539 bp and contained 132–134 genes, consisting of 86–88 coding genes, 38 transfer RNA genes and eight ribosomal RNA genes. Moreover, the characteristics of the long repeats sequences and simple sequence repeats (SSRs) were detected. In addition, we conducted phylogenomic analyses of the Zingibereae and related taxa with plastomes data from additional 32 species from Genbank. Our results confirmed that Stahlianthus is closely related to Curcuma, supporting the idea of merging it into Curcuma. Kaempferia, Boesenbergia and Zingiber were confirmed as close relatives and grouped together as the Kaempferia group. Pyrgophyllum is not allied with the Curcuma clade but instead is Citation: Liang, H.; Chen, J.