High-Resolution Genome-Wide Allelotype Analysis Identifies Loss Of
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Risk Factors for Gliomas and Meningiomas in Males in Los Angeles County1
[CANCER RESEARCH 49, 6137-6143. November 1, 1989] Risk Factors for Gliomas and Meningiomas in Males in Los Angeles County1 Susan Preston-Martin,2 Wendy Mack, and Brian E. Henderson Department of Preventive Medicine, University of Southern California School of Medicine, Los Angeles, California 90033 ABSTRACT views with proxy respondents, we were unable to include a large proportion of otherwise eligible cases because they were deceased or Detailed job histories and information about other suspected risk were too ill or impaired to participate in an interview. The Los Angeles factors were obtained during interviews with 272 men aged 25-69 with a County Cancer Surveillance Program identified the cases (26). All primary brain tumor first diagnosed during 1980-1984 and with 272 diagnoses had been microscopically confirmed. individually matched neighbor controls. Separate analyses were con A total of 478 patients were identified. The hospital and attending ducted for the 202 glioma pairs and the 70 meningioma pairs. Meningi- physician granted us permission to contact 396 (83%) patients. We oma, but not glioma, was related to having a serious head injury 20 or were unable to locate 22 patients, 38 chose not to participate, and 60 more years before diagnosis (odds ratio (OR) = 2.3; 95% confidence were aphasie or too ill to complete the interview. We interviewed 277 interval (CI) = 1.1-5.4), and a clear dose-response effect was observed patients (74% of the 374 patients contacted about the study or 58% of relating meningioma risk to number of serious head injuries (/' for trend the initial 478 patients). -

Original Article Outcome of Supratentorial Intraaxial Extra Ventricular Primary Pediatric Brain Tumors
Original Article Outcome of supratentorial intraaxial extra ventricular primary pediatric brain tumors: A prospective study Mohana Rao Patibandla, Suchanda Bhattacharjee1, Megha S. Uppin2, Aniruddh Kumar Purohit1 Department of Neurosurgery, Krishna Institute of Medical Sciences Secunderabad, Departments of 1Neurosurgery and 2Pathology, Nizam’s Institute of Medical Sciences, Hyderabad, Andhra Pradesh, India Address for correspondence: Dr. Mohana Rao Patibandla, Department of Neurosurgery, University of Colorado Denver, 13123, E 16th Ave, Aurora, CO 80045, USA. Email: [email protected] ABSTRACT Introduction: Tumors of the central nervous system (CNS) are the second most frequent malignancy of childhood and the most common solid tumor in this age group. CNS tumors represent approximately 17% of all malignancies in the pediatric age range, including adolescents. Glial neoplasms in children account for up to 60% of supratentorial intraaxial tumors. Their histological distribution and prognostic features differ from that of adults. Aims and Objectives: To study clinical and pathological characteristics, and to analyze the outcome using the Engel’s classification for seizures, Karnofsky’s score during the available follow‑up period of minimum 1 year following the surgical and adjuvant therapy of supratentorial intraaxial extraventricular primary pediatric (SIEPP) brain tumors in children equal or less than 18 years. Materials and Methods: The study design is a prospective study done in NIMS from October 2008 to January 2012. All the patients less than 18 years of age operated for SIEPP brain tumors proven histopathologically were included in the study. All the patients with recurrent or residual primary tumors or secondaries were excluded from the study. Post operative CT or magnetic resonance imaging (MRI) is done following surgery. -

Charts Chart 1: Benign and Borderline Intracranial and CNS Tumors Chart
Charts Chart 1: Benign and Borderline Intracranial and CNS Tumors Chart Glial Tumor Neuronal and Neuronal‐ Ependymomas glial Neoplasms Subependymoma Subependymal Giant (9383/1) Cell Astrocytoma(9384/1) Myyppxopapillar y Desmoplastic Infantile Ependymoma Astrocytoma (9412/1) (9394/1) Chart 1: Benign and Borderline Intracranial and CNS Tumors Chart Glial Tumor Neuronal and Neuronal‐ Ependymomas glial Neoplasms Subependymoma Subependymal Giant (9383/1) Cell Astrocytoma(9384/1) Myyppxopapillar y Desmoplastic Infantile Ependymoma Astrocytoma (9412/1) (9394/1) Use this chart to code histology. The tree is arranged Chart Instructions: Neuroepithelial in descending order. Each branch is a histology group, starting at the top (9503) with the least specific terms and descending into more specific terms. Ependymal Embryonal Pineal Choro id plexus Neuronal and mixed Neuroblastic Glial Oligodendroglial tumors tumors tumors tumors neuronal-glial tumors tumors tumors tumors Pineoblastoma Ependymoma, Choroid plexus Olfactory neuroblastoma Oligodendroglioma NOS (9391) (9362) carcinoma Ganglioglioma, anaplastic (9522) NOS (9450) Oligodendroglioma (9390) (9505 Olfactory neurocytoma Ganglioglioma, malignant (()9521) anaplastic (()9451) Anasplastic ependymoma (9505) Olfactory neuroepithlioma Oligodendroblastoma (9392) (9523) (9460) Papillary ependymoma (9393) Glioma, NOS (9380) Supratentorial primitive Atypical EdEpendymo bltblastoma MdllMedulloep ithliithelioma Medulloblastoma neuroectodermal tumor tetratoid/rhabdoid (9392) (9501) (9470) (PNET) (9473) tumor -
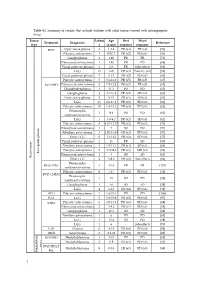
Table S1. Summary of Studies That Include Children with Solid Tumors Treated with Antiangiogenic Drugs
Table S1. Summary of studies that include children with solid tumors treated with antiangiogenic drugs. Tumor Patient Age Best Worst Treatment Diagnostic Reference type s (years) response response BVZ Optic nerve glioma 4 1.2-4 PR (x4) PR (x4) [56] Pilocytic astrocytoma 3 0.92-7 PR (x2) PD (x1) [56] Ganglioglioma 1 1.92 PR PR [56] Pilomyxoid astrocytoma 1 1.92 PR PD [68] Visual pathway glioma 1 2.6 PR Side effects [66] LGG 15 1-20 CR (x3) Toxicity (x1) [58] Visual pathway glioma 3 6-13 PR (x2) PD (x1) [60] Pilocytic astrocytoma 5 3.1-11.2 PR (x5) PR (x5) [65] BVZ+IRO Pilomyxoid astrocytoma 2 7.9-12.2 PR (x2) PR (x2) [65] Oligodendroglioma 1 11.1 PD PD [65] Ganglioglioma 3 4.1-16.8 PR (x2) SD (x1) [65] Optic nerve glioma 2 9-15 PR (x1) SD (x1) [63] LGG 35 0.6-17.6 PR (x2) PD (x8) [61] Pilocytic astrocytoma 10 1.8-15.3 PR (x4) PD (x1) [62] Pleomorphic 1 9.4 PD PD [62] xanthoastrocytoma LGG 5 3.9-9.2 PR (x2) SD (x3) [62] Pilocytic astrocytoma 4 4.67-12.17 PR (x2) PD (x3) [57] Pilomyxoid astrocytoma 1 3 SD PD [57] Fibrillary astrocytoma 3 1.92-11.08 CR (x1) PD (x3) [57] Other LGG 6 1-13.42 PR (x2) PD (x6) [57] Visual pathway glioma 1 11 PR SD [60] Fibrillary astrocytoma 3 1.5-11.1 CR (x1) SD (x1) [64] Pilocytic astrocytoma 2 3.75-9.8 PR (x1) MR (x1) [64] Low-grade glioma Pilomyxoid astrocytoma 1 3 SD SD [64] Brain tumor Brain tumor Other LGG 4 3-9.6 PR (x2) Side effects [64] Pleomorphic BVZ+TMZ 1 13.4 PR PR [153] xanthoastrocytoma Pilocytic astrocytoma 9 5-17 PR (x3) PD (x3) [59] BVZ+CHEM Pleomorphic 1 18 SD PD [59] xanthoastrocytoma Ganglioglioma -

Losses of Chromosomal Arms 1P and 19Q in the Diagnosis of Oligodendroglioma
Losses of Chromosomal Arms 1p and 19q in the Diagnosis of Oligodendroglioma. A Study of Paraffin- Embedded Sections Peter C. Burger, M.D., A. Yuriko Minn, M.S., Justin S. Smith, M.D., Ph.D., Thomas J. Borell, B.S., Anne E. Jedlicka, M.S., Brenda K. Huntley, B.S., Patricia T. Goldthwaite, M.S., Robert B. Jenkins, M.D., Ph. D., Burt G. Feuerstein, M.D., Ph.D. The Departments of Pathology (PCB, PTG) and Anesthesiology and Critical Care Medicine (AEJ), Johns Hopkins University School of Medicine, Baltimore, Maryland; Department of Pathology and Laboratory Medicine, Mayo Clinic (JSS, TJB, BKH, RBJ), Rochester, Minnesota; and Departments of Laboratory Medicine and Neurosurgery and Brain Tumor Research Center, University of California at San Francisco School of Medicine (BGF, AYM), San Francisco, California It is the impression of some pathologists that many Comparative genomic hybridization (CGH), fluores- gliomas presently designated as oligodendrogliomas cence in situ hybridization (FISH), polymerase chain would have been classified in the past as astrocyto- reaction–based microsatellite analysis, and p53 se- mas with little thought about the possibility of an quencing were performed in paraffin-embedded ma- oligodendroglial component. It also appears that the terial from 18 oligodendrogliomas and histologically relative incidence of the diagnoses of oligodendrogli- similar astrocytomas. The study was undertaken be- oma and astrocytomas varies widely from institution cause of evidence that concurrent loss of both the 1p to institution, suggesting that diagnostic criteria dif- and 19q chromosome arms is a specific marker for fer. In some laboratories, even minor or focal nuclear oligodendrogliomas. Of the six lesions with a review roundness or perinuclear haloes are considered to be diagnosis of oligodendroglioma, all had the predicted evidence of oligodendroglial differentiation, whereas loss of 1p and 19q seen by CGH, FISH, and polymerase pathologists elsewhere require the more classical fea- chain reaction. -

Brain and Spine Tumors
Brain and Spine Tumors Andrew J. Fabiano, MD FAANS Associate Professor of Neurosurgery Roswell Park Cancer Institute SUNY at Buffalo School of Medicine Brain Tumors Brain Tumor Basics Types of Tumors Cases Brain Tumors Skull is a fixed space Symptoms develop due to compression of normal brain Brain Tumors Brain Tumors Inflammation/Edema occurs in the surrounding normal brain Brain Tumors Tumors cause edema and irritation of normal brain Breakdown of BBB Corticosteroids for edema Anti-epileptics to prevent seizures Corticosteroids Dexamethasone traditionally used Reduces vasogenic edema GI prophylaxis Steroids Multiple side effects: Diabetes Myopathy Infection LE edema Weight gain Wound issues Anti-Epileptic Drugs Used for cortical lesions Not required for cerebellar lesions Dilantin – requires monitoring Keppra Tumor Types Gliomas Meningiomas Metastatic Tumors Pituitary Tumors Gliomas Arise from native cells within the brain Gliomas WHO I – Pilocytic Astrocytoma WHO II – Fibrillary Astrocytoma WHO III – Anaplastic Astrocytoma WHO IV – Glioblastoma Multiforme Gliomas – WHO I Gliomas – WHO II & III WHO IV - GBM Glioblastoma Multiforme Most common primary brain tumor in adults Incidence 3 per 100,000 Average survival from diagnosis ~ 13 months Young age, High Karnofsky score associated with increased survival Glioblastoma Start steroids and anti-epileptics Glioblastoma Gliomas - Treatment Surgery Biopsy External Beam XRT Chemotherapy (Temodar) Glioblastoma Survival Related to Extent of Resection Glioblastoma Typical IMRT course is Monday-Friday -

Conversion of a Gemistocytic Astrocytoma to a Fibrillary Astrocytoma After Temozolomide and Radiation Therapy
Open Access Case Report DOI: 10.7759/cureus.219 Conversion of a Gemistocytic Astrocytoma to a Fibrillary Astrocytoma after Temozolomide and Radiation Therapy Brandon C. Gabel, Mary Goolsby, Lawrence Hansen, Bob Carter, Clark Chen 1. Corresponding author: Brandon C. Gabel, [email protected] Abstract In most instances, gemistocytic astrocytoma progresses to anaplastic astrocytoma at the time of recurrence. Here, we report an unusual case where a gemistocytic astrocytoma converted to a more benign fibrillary histology after temozolomide/radiation therapy. We present the case of a 26-year-old woman with a history of fibrillary astrocytoma that was followed over a span of eight years and underwent three craniotomies. The pathology from the first craniotomy revealed a fibrillary astrocytoma. The pathology from the second craniotomy (one year after the first resection) revealed findings consistent with a gemistocytic astrocytoma. The pathology from a third craniotomy (seven years after the second resection) revealed a fibrillary astrocytoma without a significant gemistocytic component. This is the first documented case of a gemistocytic astrocytoma converting to a more benign fibrillary pathology at the time of recurrence. We speculate that the phenomenon is the result of the inherently heterogeneous astrocytic subpopulations that were differentially sensitive to chemo/radiation. Categories: Neurology, Neurosurgery, Oncology Keywords: gemistocytic, fibrillary, astrocytoma, temozolomide, radiation, glioma Introduction It is a commonly accepted truism in oncology that the grade and aggressiveness of a tumor increases at the time of recurrence. As an example of this phenomenon, recurrent gemistocytic astrocytoma nearly always exhibit less differentiated or anaplastic histology [1-2]. We present an interesting case of a gemistocytic astrocytoma that converted to a more benign fibrillary histology at the time of recurrence. -

Low-Grade Gliomas 26
C H A P T E R Low-Grade Gliomas 26 Paul D. Brown, Michael D. Chan, Edward G. Shaw, and Martin J. van den Bent EPIDEMIOLOGIC AND ETIOLOGIC FINDINGS PRIMARY THERAPY The estimated annual incidence of low-grade gliomas in the Maximum surgical resection is associated with a more favor- United States is 2000 cases. Etiologic factors are unknown. able outcome. These tumors are seen with increased frequency in patients In pediatric low-grade gliomas, observation is recom- with types 1 and 2 neurofibromatosis. mended after gross total resection. After subtotal resection or biopsy, treatment (radiation therapy, chemotherapy) is usually PATHOLOGIC FINDINGS deferred until symptomatic or imaging progression. The most common pathologic types include pilocytic astrocy- In adults, after gross total resection of a pilocytic astrocy- toma (World Health Organization [WHO] grade I), diffuse toma, observation is recommended. After gross total or subto- fibrillary astrocytoma, oligoastrocytoma, and oligodendrogli- tal resection (or biopsy) of a low-grade glioma, irradiation oma (WHO grade II). may be given postoperatively or deferred until progression occurs. BIOLOGIC CHARACTERISTICS The recommended radiotherapy (RT) dose is 45 Gy to Combined 1p and 19q deletions are associated with a superior 54 Gy in 25 fractions to 30 fractions, treating the MRI-defined outcome and are most common in oligodendrogliomas. TP53 tumor volume with a 2-cm margin. mutations are more common in diffuse astrocytomas and are Early results from one large randomized clinical trial mutually exclusive from 1p/19q co-deletions. IDH1 mutations suggest disease and outcomes are similar for either temozolo- occur in the vast majority of low-grade gliomas and are found mide or RT for 1p/19q-deleted tumors although RT may have both in tumors with TP53 mutations and in tumors with a superior PFS for 1p-intact tumors; however, the period of 1p/19q codeletions. -
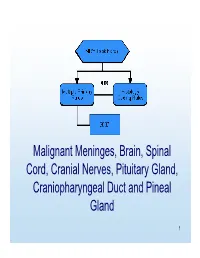
MP/H Rules Presentation
Malignant Meninges, Brain, Spinal Cord, Cranial Nerves, Pituitary Gland, Craniopharyngeal Duct and Pineal Gland 1 Equivalent Terms, Definitions, Charts and Illustrations • Benign and borderline intracranial and CNS tumors have a separate set of rules. 2 Equivalent Terms, Definitions, Charts and Illustrations • PNET (Primitive neuroectodermal tumor) – Central PNET – Supratentorial PNET • pPNET –not brain primary 3 Chart 1 Neuroepithelial Brain CNS • WHO Classification of Tumors of the brain and central nervous system • Not complete listing 4 Chart Instructions: Use this chart to code histology. The tree is arranged Neuroepithelial in descending order. Each branch is a histology group, starting at the top (9503) with the least specific terms and descending into more specific terms. Embryonal Ependymal Pineal Choroid plexus Neuronal and mixed Neuroblastic Glial Oligodendroglial tumors tumors tumors tumors neuronal-glial tumors tumors tumors tumors Ependymoma, Pineoblastoma Choroid plexus Olfactory neuroblastoma Oligodendroglioma NOS (9391) (9362) carcinoma Ganglioglioma, anaplastic (9522) NOS (9450) (9390) (9505 Olfactory neurocytoma Oligodendroglioma Ganglioglioma, malignant (9521) anaplastic (9451) Anasplastic ependymoma (9505) Olfactory neuroepithlioma Oligodendroblastoma (9392) (9523) (9460) Papillary ependymoma (9393) Glioma, NOS (9380) Atypical Ependymoblastoma Medulloepithelioma Medulloblastoma Supratentorial primitive tetratoid/rhabdoid (9392) (9501) (9470) neuroectodermal tumor tumor (9508) (PNET) (9473) Teratoid Demoplastic (9471) -

Pediatric Supratentorial Intraventricular Tumors
Neurosurg Focus 10 (6):Article 4, 2001, Click here to return to Table of Contents Pediatric supratentorial intraventricular tumors DANIEL Y. SUH, M.D., PH.D., AND TIMOTHY MAPSTONE, M.D. Department of Neurosurgery, Emory University School of Medicine, and the Children’s Healthcare of Atlanta-Egleston, Atlanta, Georgia A variety of mass lesions can arise within or in proximity to the ventricular system in children. These lesions are relatively uncommon, and they present a unique diagnostic and surgical challenge. The differential diagnosis is deter- mined by tumor location in the ventricular system, clinical presentation, age of the patient, and the imaging character- istics of the lesion. In this report the authors provide an introduction to and an overview of the most common pediatric supratentorial intraventricular tumors. The typical radiographic features of each tumor and location preference within the ventricular system are reviewed. Management and treatment considerations are discussed. Examination of tissue samples to obtain diagnosis is usually required for accurate treatment planning, and resection without adjuvant thera- pies is often curative. The critical management decision frequently involves determining which lesions are appropri- ate for surgical therapy. Care ful preoperative neuroimaging is extremely useful in planning surgery. Knowledge of the typical imaging characteristics of these tumors can help to determine the diagnosis with relative certainty when a tis- sue sample has not been obtained, because a small subset of these lesions can be managed expectantly. KEY WORDS • intraventricular tumor • pediatric tumor • brain lesion • hydrocephalus One tenth of all CNS neoplasms present within or in certain anatomical locations and in certain age groups.61,75, proximity to the ventricular system.85 These neoplasms 80,85,92,124,133 Table 1 provides a list of common pediatric comprise a heterogeneous group with regard to tumor type intraventricular tumors by ventricular location. -

1. Tumours of Neuroepithelial Tissue • • • • • • • • • • • • • • • •
WHO Classification of Tumors of the Central Nervous System 1. Tumours of Neuroepithelial 1.4 Ependymal tumours Tissue • Ependymoma 1.1 Astrocytic tumours • Variants: Cellular • Diffuse astrocytoma • Papillary • Variants: Fibrillary astrocytoma • Clear cell • Protoplasmic astrocytoma • Tanycytic • Gemistocytic astrocytoma • Anaplastic ependymoma • Anaplastic astrocytoma • Myxopapillary ependymoma • Glioblastoma • Subependymoma • Variants: Giant cell glioblastoma 1.5 Choroid plexus tumours • Gliosarcoma • Choroid plexus papilloma • Pilocytic astrocytoma • Choroid plexus carcinoma • Pleomorphic xanthoastrocytoma 1.6 Glial tumours of uncertain origin • Subependymal giant cell astrocytoma • Astroblastoma 1.2 Oligodendroglial tumours • Gliomatosis cerebri • Oligodendroglioma • Chordoid glioma of the 3rd ventricle • Anaplastic oligodendroglioma 1.7 Neuronal and mixed neuronalglial tumours 1.3 Mixed gliomas • Gangliocytoma • Oligoastrocytoma • Dysplastic gangliocytoma of cerebellum (Lhermitte-Duclos) • Anaplastic oligoastrocytoma • Desmoplastic infantile astrocytoma/ganglioglioma • Oligodendroglioma • Dysembryoplastic neuroepithelial tumour • Anaplastic oligodendroglioma • Ganglioglioma Page 1 of 4 WHO Classification of Tumors of the Central Nervous System • Anaplastic ganglioglioma • tumour (PNET) • Central neurocytoma • Variants: Neuroblastoma • Cerebellar liponeurocytoma • Ganglioneuroblastoma • Paraganglioma of the filum terminale • Atypical teratoid/rhabdoid tumour 1.8 Neuroblastic tumours • Olfactory neuroblastoma (Aesthesioneuroblastoma) -

Optic Nerve Glioma in Mice Requires Astrocyte Nf1 Gene Inactivation and Nf1 Brain Heterozygosity
[CANCER RESEARCH 63, 8573–8577, December 15, 2003] Advances in Brief Optic Nerve Glioma in Mice Requires Astrocyte Nf1 Gene Inactivation and Nf1 Brain Heterozygosity M. Livia Bajenaru,1 M. Rosario Hernandez,2 Arie Perry,3 Yuan Zhu,6 Luis F. Parada,6 Joel R. Garbow,4,5 and David H. Gutmann1 Departments of 1Neurology, 2Ophthalmology, 3Pathology, 4Radiology, and 5Chemistry, Washington University School of Medicine, St. Louis, Missouri, and 6Center for Developmental Biology and Kent Waldrep Foundation Center for Basic Research on Nerve Growth and Regeneration, University of Texas Southwestern Medical Center, Dallas, Texas Abstract bryonic day 14 using Cre/LoxP technology (11). Glial fibrillary acidic protein (GFAP) Cre; Nf1flox/flox mice are viable and fertile and exhibit Whereas biallelic neurofibromatosis 1 (NF1) inactivation is observed in increased numbers of brain and optic nerve astrocytes, but they do not NF1-associated gliomas, astrocyte-restricted Nf1 conditional knockout develop gliomas. The absence of glioma formation in these mice, even mice do not develop gliomas. These observations suggest that NF1 glioma formation requires additional cellular or genetic conditions. To determine after 20 months of age, suggests that additional cellular or genetic the effect of an Nf1 heterozygous brain environment on NF1 glioma events are necessary for glioma tumorigenesis in the setting of NF1. -formation, we generated Nf1؉/؊ mice lacking Nf1 expression in astro- Because patients with NF1 are heterozygous for a germ-line inacti cytes. In contrast to astrocyte-restricted Nf1 conditional knockout mice, vating NF1 mutation, and Nf1 mutant mice develop neurofibroma, Nf1؉/؊ mice lacking Nf1 in astrocytes develop optic nerve gliomas.