The Misclassification of Diffuse Gliomas: Rates and Outcomes J
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Risk Factors for Gliomas and Meningiomas in Males in Los Angeles County1
[CANCER RESEARCH 49, 6137-6143. November 1, 1989] Risk Factors for Gliomas and Meningiomas in Males in Los Angeles County1 Susan Preston-Martin,2 Wendy Mack, and Brian E. Henderson Department of Preventive Medicine, University of Southern California School of Medicine, Los Angeles, California 90033 ABSTRACT views with proxy respondents, we were unable to include a large proportion of otherwise eligible cases because they were deceased or Detailed job histories and information about other suspected risk were too ill or impaired to participate in an interview. The Los Angeles factors were obtained during interviews with 272 men aged 25-69 with a County Cancer Surveillance Program identified the cases (26). All primary brain tumor first diagnosed during 1980-1984 and with 272 diagnoses had been microscopically confirmed. individually matched neighbor controls. Separate analyses were con A total of 478 patients were identified. The hospital and attending ducted for the 202 glioma pairs and the 70 meningioma pairs. Meningi- physician granted us permission to contact 396 (83%) patients. We oma, but not glioma, was related to having a serious head injury 20 or were unable to locate 22 patients, 38 chose not to participate, and 60 more years before diagnosis (odds ratio (OR) = 2.3; 95% confidence were aphasie or too ill to complete the interview. We interviewed 277 interval (CI) = 1.1-5.4), and a clear dose-response effect was observed patients (74% of the 374 patients contacted about the study or 58% of relating meningioma risk to number of serious head injuries (/' for trend the initial 478 patients). -

Original Article Outcome of Supratentorial Intraaxial Extra Ventricular Primary Pediatric Brain Tumors
Original Article Outcome of supratentorial intraaxial extra ventricular primary pediatric brain tumors: A prospective study Mohana Rao Patibandla, Suchanda Bhattacharjee1, Megha S. Uppin2, Aniruddh Kumar Purohit1 Department of Neurosurgery, Krishna Institute of Medical Sciences Secunderabad, Departments of 1Neurosurgery and 2Pathology, Nizam’s Institute of Medical Sciences, Hyderabad, Andhra Pradesh, India Address for correspondence: Dr. Mohana Rao Patibandla, Department of Neurosurgery, University of Colorado Denver, 13123, E 16th Ave, Aurora, CO 80045, USA. Email: [email protected] ABSTRACT Introduction: Tumors of the central nervous system (CNS) are the second most frequent malignancy of childhood and the most common solid tumor in this age group. CNS tumors represent approximately 17% of all malignancies in the pediatric age range, including adolescents. Glial neoplasms in children account for up to 60% of supratentorial intraaxial tumors. Their histological distribution and prognostic features differ from that of adults. Aims and Objectives: To study clinical and pathological characteristics, and to analyze the outcome using the Engel’s classification for seizures, Karnofsky’s score during the available follow‑up period of minimum 1 year following the surgical and adjuvant therapy of supratentorial intraaxial extraventricular primary pediatric (SIEPP) brain tumors in children equal or less than 18 years. Materials and Methods: The study design is a prospective study done in NIMS from October 2008 to January 2012. All the patients less than 18 years of age operated for SIEPP brain tumors proven histopathologically were included in the study. All the patients with recurrent or residual primary tumors or secondaries were excluded from the study. Post operative CT or magnetic resonance imaging (MRI) is done following surgery. -

Charts Chart 1: Benign and Borderline Intracranial and CNS Tumors Chart
Charts Chart 1: Benign and Borderline Intracranial and CNS Tumors Chart Glial Tumor Neuronal and Neuronal‐ Ependymomas glial Neoplasms Subependymoma Subependymal Giant (9383/1) Cell Astrocytoma(9384/1) Myyppxopapillar y Desmoplastic Infantile Ependymoma Astrocytoma (9412/1) (9394/1) Chart 1: Benign and Borderline Intracranial and CNS Tumors Chart Glial Tumor Neuronal and Neuronal‐ Ependymomas glial Neoplasms Subependymoma Subependymal Giant (9383/1) Cell Astrocytoma(9384/1) Myyppxopapillar y Desmoplastic Infantile Ependymoma Astrocytoma (9412/1) (9394/1) Use this chart to code histology. The tree is arranged Chart Instructions: Neuroepithelial in descending order. Each branch is a histology group, starting at the top (9503) with the least specific terms and descending into more specific terms. Ependymal Embryonal Pineal Choro id plexus Neuronal and mixed Neuroblastic Glial Oligodendroglial tumors tumors tumors tumors neuronal-glial tumors tumors tumors tumors Pineoblastoma Ependymoma, Choroid plexus Olfactory neuroblastoma Oligodendroglioma NOS (9391) (9362) carcinoma Ganglioglioma, anaplastic (9522) NOS (9450) Oligodendroglioma (9390) (9505 Olfactory neurocytoma Ganglioglioma, malignant (()9521) anaplastic (()9451) Anasplastic ependymoma (9505) Olfactory neuroepithlioma Oligodendroblastoma (9392) (9523) (9460) Papillary ependymoma (9393) Glioma, NOS (9380) Supratentorial primitive Atypical EdEpendymo bltblastoma MdllMedulloep ithliithelioma Medulloblastoma neuroectodermal tumor tetratoid/rhabdoid (9392) (9501) (9470) (PNET) (9473) tumor -

Paraganglioma (PGL) Tumors in Patients with Succinate Dehydrogenase-Related PCC–PGL Syndromes: a Clinicopathological and Molecular Analysis
T G Papathomas and others Non-PCC/PGL tumors in the SDH 170:1 1–12 Clinical Study deficiency Non-pheochromocytoma (PCC)/paraganglioma (PGL) tumors in patients with succinate dehydrogenase-related PCC–PGL syndromes: a clinicopathological and molecular analysis Thomas G Papathomas1, Jose Gaal1, Eleonora P M Corssmit2, Lindsey Oudijk1, Esther Korpershoek1, Ketil Heimdal3, Jean-Pierre Bayley4, Hans Morreau5, Marieke van Dooren6, Konstantinos Papaspyrou7, Thomas Schreiner8, Torsten Hansen9, Per Arne Andresen10, David F Restuccia1, Ingrid van Kessel6, Geert J L H van Leenders1, Johan M Kros1, Leendert H J Looijenga1, Leo J Hofland11, Wolf Mann7, Francien H van Nederveen12, Ozgur Mete13,14, Sylvia L Asa13,14, Ronald R de Krijger1,15 and Winand N M Dinjens1 1Department of Pathology, Josephine Nefkens Institute, Erasmus MC, University Medical Center, PO Box 2040, 3000 CA Rotterdam, The Netherlands, 2Department of Endocrinology, Leiden University Medical Center, Leiden,The Netherlands, 3Section for Clinical Genetics, Department of Medical Genetics, Oslo University Hospital, Oslo, Norway, 4Department of Human and Clinical Genetics, Leiden University Medical Center, Leiden, The Netherlands, 5Department of Pathology, Leiden University Medical Center, Leiden, The Netherlands, 6Department of Clinical Genetics, Erasmus MC, University Medical Center, Rotterdam, The Netherlands, 7Department of Otorhinolaryngology, Head and Neck Surgery, University Medical Center of the Johannes Gutenberg University Mainz, Mainz, Germany, 8Section for Specialized Endocrinology, -

Central Nervous System Tumors General ~1% of Tumors in Adults, but ~25% of Malignancies in Children (Only 2Nd to Leukemia)
Last updated: 3/4/2021 Prepared by Kurt Schaberg Central Nervous System Tumors General ~1% of tumors in adults, but ~25% of malignancies in children (only 2nd to leukemia). Significant increase in incidence in primary brain tumors in elderly. Metastases to the brain far outnumber primary CNS tumors→ multiple cerebral tumors. One can develop a very good DDX by just location, age, and imaging. Differential Diagnosis by clinical information: Location Pediatric/Young Adult Older Adult Cerebral/ Ganglioglioma, DNET, PXA, Glioblastoma Multiforme (GBM) Supratentorial Ependymoma, AT/RT Infiltrating Astrocytoma (grades II-III), CNS Embryonal Neoplasms Oligodendroglioma, Metastases, Lymphoma, Infection Cerebellar/ PA, Medulloblastoma, Ependymoma, Metastases, Hemangioblastoma, Infratentorial/ Choroid plexus papilloma, AT/RT Choroid plexus papilloma, Subependymoma Fourth ventricle Brainstem PA, DMG Astrocytoma, Glioblastoma, DMG, Metastases Spinal cord Ependymoma, PA, DMG, MPE, Drop Ependymoma, Astrocytoma, DMG, MPE (filum), (intramedullary) metastases Paraganglioma (filum), Spinal cord Meningioma, Schwannoma, Schwannoma, Meningioma, (extramedullary) Metastases, Melanocytoma/melanoma Melanocytoma/melanoma, MPNST Spinal cord Bone tumor, Meningioma, Abscess, Herniated disk, Lymphoma, Abscess, (extradural) Vascular malformation, Metastases, Extra-axial/Dural/ Leukemia/lymphoma, Ewing Sarcoma, Meningioma, SFT, Metastases, Lymphoma, Leptomeningeal Rhabdomyosarcoma, Disseminated medulloblastoma, DLGNT, Sellar/infundibular Pituitary adenoma, Pituitary adenoma, -

Ambient Mass Spectrometry for the Intraoperative Molecular Diagnosis of Human Brain Tumors
Ambient mass spectrometry for the intraoperative molecular diagnosis of human brain tumors Livia S. Eberlina, Isaiah Nortonb, Daniel Orringerb, Ian F. Dunnb, Xiaohui Liub, Jennifer L. Ideb, Alan K. Jarmuscha, Keith L. Ligonc, Ferenc A. Joleszd, Alexandra J. Golbyb,d, Sandro Santagatac, Nathalie Y. R. Agarb,d,1, and R. Graham Cooksa,1 aDepartment of Chemistry and Center for Analytical Instrumentation Development, Purdue University, West Lafayette, IN 47907; and Departments of bNeurosurgery, cPathology, and dRadiology, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02115 Edited by Jack Halpern, The University of Chicago, Chicago, IL, and approved December 5, 2012 (received for review September 11, 2012) The main goal of brain tumor surgery is to maximize tumor resection at Brigham and Women’s Hospital (BWH), created an opportu- while preserving brain function. However, existing imaging and nity for collecting information about the extent of tumor resection surgical techniques do not offer the molecular information needed during surgery (5, 6). Although brain tumor resection typically to delineate tumor boundaries. We have developed a system to requires multiple hours, intraoperative MRI can be completed rapidly analyze and classify brain tumors based on lipid information and information evaluated within an hour. However, MRI has acquired by desorption electrospray ionization mass spectrometry limited ability to distinguish residual tumor from surrounding (DESI-MS). In this study, a classifier was built to discriminate gliomas normal brain (9). In consequence, there is a need for more de- and meningiomas based on 36 glioma and 19 meningioma samples. tailed molecular information to be acquired on a timescale closer The classifier was tested and results were validated for intraoper- to real time than can be supplied by MRI. -

Cerebellar Anaplastic Astrocytoma in Adult Patients: 15 Consecutive Cases from a Single Institution and Literature Review
medRxiv preprint doi: https://doi.org/10.1101/2020.09.09.20188938; this version posted September 14, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. All rights reserved. No reuse allowed without permission. Cerebellar anaplastic astrocytoma in adult patients: 15 consecutive cases from a single institution and literature review Artem Belyaev1, Dmitry Usachev1, Marina Ryzhova1, Gleb Gulida1, Vasilisa Skvortsova2,3, Igor Pronin1, Grigory Kobiakov1. 1 Burdenko Neurosurgery Center, 4th Tverskaya-Yamskaya, 16, Moscow, 125047, Russia 2 Wellcome Trust Centre for Neuroimaging, University College London, London, United Kingdom; 3 Max Planck University College London Centre for Computational Psychiatry and Ageing Research, London, United Kingdom; Abstract: Adult cerebellar anaplastic astrocytomas (cAA) are rare entities and their clinical and genetic appearances are still ill defined. Previously, malignant gliomas of the cerebellum were combined and reviewed together (cAA and cerebellar glioblastomas (cGB), that could have possibly affected overall results. We present characteristics of 15 adult patients with cAA and compared them to a series of 45 patients with a supratentorial AA (sAA). The mean age at cAA diagnosis was 39.3 years (range 19-72). A history of neurofibromatosis type I was noted in 1 patient (6.7%). An IDH-1 mutation was identified in 6/15 cases and a methylated MGMT promoter in 5/15 cases. Patients in study and control groups were matched in age, sex and IDH- 1 mutation status. Patients in a study group tended to have a more frequent multifocal presentation at diagnosis (13% vs. -
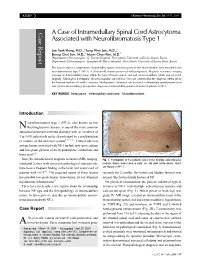
A Case of Intramedullary Spinal Cord Astrocytoma Associated with Neurofibromatosis Type 1
KISEP J Korean Neurosurg Soc 36 : 69-71, 2004 Case Report A Case of Intramedullary Spinal Cord Astrocytoma Associated with Neurofibromatosis Type 1 Jae Taek Hong, M.D.,1 Sang Won Lee, M.D.,1 Byung Chul Son, M.D.,1 Moon Chan Kim, M.D.2 Department of Neurosurgery,1 St. Vincent Hospital, The Catholic University of Korea, Suwon, Korea Department of Neurosurgery,2 Kangnam St. Mary's Hospital, The Catholic University of Korea, Seoul, Korea The authors report a symptomatic intramedullary spinal cord astrocytoma in the thoracolumbar area associated with neurofibromatosis type 1 (NF-1). A 38-year-old woman presented with paraparesis. Magnetic resonance imaging revealed an intramedullary lesion within the lower thoracic spinal cord and conus medullaris, which was removed surgically. Pathological investigation showed anaplastic astrocytoma. This case confirms that the diagnosis criteria set by the National Institute of Health Consensus Development Conference can be useful to differentiate ependymoma from astrocytoma when making a preoperative diagnosis of intramedullary spinal cord tumor in patients of NF-1. KEY WORDS : Astrocytoma·Intramedullary cord tumor·Neurofibromatosis. Introduction eurofibromatosis type 1 (NF-1), also known as von N Recklinghausen's disease, is one of the most common autosomal dominant inherited disorders with an incidence of 1 in 3,000 individuals and is characterized by a predisposition to tumors of the nervous system5,6,12,16). Central nervous system lesions associated with NF-1 include optic nerve glioma and low-grade gliomas of the hypothalamus, cerebellum and brain stem6,10). Since the introduction of magnetic resonance(MR) imaging, Fig. 1. Photograph of the patient's back shows multiple subcutaneous incidental lesions with uncertain pathological characteristic nodules (black arrow) and a cafe-au-lait spot (white arrow), which have been a frequent finding in the brain and spinal cord of are typical of NF-1. -

Central Neurocytoma SNP Array Analyses, Subtel FISH, and Review
Pathology - Research and Practice 215 (2019) 152397 Contents lists available at ScienceDirect Pathology - Research and Practice journal homepage: www.elsevier.com/locate/prp Case report Central neurocytoma: SNP array analyses, subtel FISH, and review of the T literature Caroline Sandera,1, Marco Wallenborna,b,1, Vivian Pascal Brandtb, Peter Ahnertc, Vera Reuscheld, ⁎ Christan Eisenlöffele, Wolfgang Kruppa, Jürgen Meixensbergera, Heidrun Hollandb, a Dept. of Neurosurgery, University of Leipzig, Liebigstraße 26, 04103 Leipzig, Germany b Saxonian Incubator for Clinical Translation, University of Leipzig, Philipp-Rosenthal Str. 55, 04103 Leipzig, Germany c Institute for Medical Informatics, Statistics and Epidemiology, University of Leipzig, Haertelstraße 16-18, 04107 Leipzig, Germany d Dept. of Neuroradiology, University of Leipzig, Liebigstraße 22a, 04103 Leipzig, Germany e Dept. of Neuropathology, University of Leipzig, Liebigstraße 26, 04103 Leipzig, Germany ARTICLE INFO ABSTRACT Keywords: The central neurocytoma (CN) is a rare brain tumor with a frequency of 0.1-0.5% of all brain tumors. According Central neurocytoma to the World Health Organization classification, it is a benign grade II tumor with good prognosis. However, Cytogenetics some CN occur as histologically “atypical” variant, combined with increasing proliferation and poor clinical SNP array outcome. Detailed genetic knowledge could be helpful to characterize a potential atypical behavior in CN. Only FISH few publications on genetics of CN exist in the literature. Therefore, we performed cytogenetic analysis of an intraventricular neurocytoma WHO grade II in a 39-year-old male patient by use of genome-wide high-density single nucleotide polymorphism array (SNP array) and subtelomere FISH. Applying these techniques, we could detect known chromosomal aberrations and identified six not previously described chromosomal aberrations, gains of 1p36.33-p36.31, 2q37.1-q37.3, 6q27, 12p13.33-p13.31, 20q13.31-q13.33, and loss of 19p13.3-p12. -

Inhibition of Mir-1193 Leads to Synthetic Lethality in Glioblastoma
Zhang et al. Cell Death and Disease (2020) 11:602 https://doi.org/10.1038/s41419-020-02812-3 Cell Death & Disease ARTICLE Open Access Inhibition of miR-1193 leads to synthetic lethality in glioblastoma multiforme cells deficient of DNA-PKcs Jing Zhang1,LiJing1,SubeeTan2, Er-Ming Zeng3, Yingbo Lin4, Lingfeng He 1, Zhigang Hu 1, Jianping Liu1 and Zhigang Guo1 Abstract Glioblastoma multiforme (GBM) is the most malignant primary brain tumor and has the highest mortality rate among cancers and high resistance to radiation and cytotoxic chemotherapy. Although some targeted therapies can partially inhibit oncogenic mutation-driven proliferation of GBM cells, therapies harnessing synthetic lethality are ‘coincidental’ treatments with high effectiveness in cancers with gene mutations, such as GBM, which frequently exhibits DNA-PKcs mutation. By implementing a highly efficient high-throughput screening (HTS) platform using an in-house-constructed genome-wide human microRNA inhibitor library, we demonstrated that miR-1193 inhibition sensitized GBM tumor cells with DNA-PKcs deficiency. Furthermore, we found that miR-1193 directly targets YY1AP1, leading to subsequent inhibition of FEN1, an important factor in DNA damage repair. Inhibition of miR-1193 resulted in accumulation of DNA double-strand breaks and thus increased genomic instability. RPA-coated ssDNA structures enhanced ATR checkpoint kinase activity, subsequently activating the CHK1/p53/apoptosis axis. These data provide a preclinical theory for the application of miR-1193 inhibition as a potential synthetic lethal approach targeting GBM cancer cells with DNA-PKcs fi 1234567890():,; 1234567890():,; 1234567890():,; 1234567890():,; de ciency. Introduction In response to DNA damage, cells activate the DNA Glioblastoma multiforme (GBM), exhibits highly damage response (DDR) network, allowing DNA repair aggressive invasion, a high mortality rate, and high resis- through the regulation of cell-cycle progression, DNA tance to radiation and cytotoxic chemotherapy, and thus damage repair or apoptosis4. -

A Case of Mushroom‑Shaped Anaplastic Oligodendroglioma Resembling Meningioma and Arteriovenous Malformation: Inadequacies of Diagnostic Imaging
EXPERIMENTAL AND THERAPEUTIC MEDICINE 10: 1499-1502, 2015 A case of mushroom‑shaped anaplastic oligodendroglioma resembling meningioma and arteriovenous malformation: Inadequacies of diagnostic imaging YAOLING LIU1,2, KANG YANG1, XU SUN1, XINYU LI1, MINGHAI WEI1, XIANG'EN SHI2, NINGWEI CHE1 and JIAN YIN1 1Department of Neurosurgery, The Second Affiliated Hospital of Dalian Medical University, Dalian, Liaoning 116044; 2Department of Neurosurgery, Affiliated Fuxing Hospital, The Capital University of Medical Sciences, Beijing 100038, P.R. China Received December 29, 2014; Accepted June 29, 2015 DOI: 10.3892/etm.2015.2676 Abstract. Magnetic resonance imaging (MRI) is the most tomas (WHO IV) (2). The median survival times of patients widely discussed and clinically employed method for the with WHO II and WHO III oligodendrogliomas are 9.8 and differential diagnosis of oligodendrogliomas; however, 3.9 years, respectively (1,2), and 6.3 and 2.8 years, respec- MRI occasionally produces unclear results that can hinder tively, if mixed with astrocytes (3,4). Surgical excision and a definitive oligodendroglioma diagnosis. The present study postoperative adjuvant radiotherapy is the traditional therapy describes the case of a 34-year-old man that suffered from for oligodendroglioma; however, studies have observed that, headache and right upper‑extremity weakness for 2 months. among intracranial tumors, anaplastic oligodendrogliomas Based on the presurgical evaluation, it was suggested that the are particularly sensitive to chemotherapy, and the prognosis patient had a World Health Organization (WHO) grade II-II of patients treated with chemotherapy is more favorable glioma, meningioma or arteriovenous malformation (AVM), than that of patients treated with radiotherapy (5‑7). -

Astrocytoma: a Hormone-Sensitive Tumor?
International Journal of Molecular Sciences Review Astrocytoma: A Hormone-Sensitive Tumor? Alex Hirtz 1, Fabien Rech 1,2,Hélène Dubois-Pot-Schneider 1 and Hélène Dumond 1,* 1 Université de Lorraine, CNRS, CRAN, F-54000 Nancy, France; [email protected] (A.H.); [email protected] (F.R.); [email protected] (H.D.-P.-S.) 2 Université de Lorraine, CHRU-Nancy, Service de Neurochirurgie, F-54000 Nancy, France * Correspondence: [email protected]; Tel.: +33-372746115 Received: 29 October 2020; Accepted: 27 November 2020; Published: 30 November 2020 Abstract: Astrocytomas and, in particular, their most severe form, glioblastoma, are the most aggressive primary brain tumors and those with the poorest vital prognosis. Standard treatment only slightly improves patient survival. Therefore, new therapies are needed. Very few risk factors have been clearly identified but many epidemiological studies have reported a higher incidence in men than women with a sex ratio of 1:4. Based on these observations, it has been proposed that the neurosteroids and especially the estrogens found in higher concentrations in women’s brains could, in part, explain this difference. Estrogens can bind to nuclear or membrane receptors and potentially stimulate many different interconnected signaling pathways. The study of these receptors is even more complex since many isoforms are produced from each estrogen receptor encoding gene through alternative promoter usage or splicing, with each of them potentially having a specific role in the cell. The purpose of this review is to discuss recent data supporting the involvement of steroids during gliomagenesis and to focus on the potential neuroprotective role as well as the mechanisms of action of estrogens in gliomas.