The New WHO Classification of Brain Tumors and Molecular Profiling in the Diagnosis of Gliomas
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Central Nervous System Cancers Panel Members Can Be Found on Page 1151
1114 NCCN David Tran, MD, PhD; Nam Tran, MD, PhD; Frank D. Vrionis, MD, MPH, PhD; Patrick Y. Wen, MD; Central Nervous Nicole McMillian, MS; and Maria Ho, PhD System Cancers Overview In 2013, an estimated 23,130 people in the United Clinical Practice Guidelines in Oncology States will be diagnosed with primary malignant brain Louis Burt Nabors, MD; Mario Ammirati, MD, MBA; and other central nervous system (CNS) neoplasms.1 Philip J. Bierman, MD; Henry Brem, MD; Nicholas Butowski, MD; These tumors will be responsible for approximately Marc C. Chamberlain, MD; Lisa M. DeAngelis, MD; 14,080 deaths. The incidence of primary brain tumors Robert A. Fenstermaker, MD; Allan Friedman, MD; Mark R. Gilbert, MD; Deneen Hesser, MSHSA, RN, OCN; has been increasing over the past 30 years, especially in Matthias Holdhoff, MD, PhD; Larry Junck, MD; elderly persons.2 Metastatic disease to the CNS occurs Ronald Lawson, MD; Jay S. Loeffler, MD; Moshe H. Maor, MD; much more frequently, with an estimated incidence ap- Paul L. Moots, MD; Tara Morrison, MD; proximately 10 times that of primary brain tumors. An Maciej M. Mrugala, MD, PhD, MPH; Herbert B. Newton, MD; Jana Portnow, MD; Jeffrey J. Raizer, MD; Lawrence Recht, MD; estimated 20% to 40% of patients with systemic cancer Dennis C. Shrieve, MD, PhD; Allen K. Sills Jr, MD; will develop brain metastases.3 Abstract Please Note Primary and metastatic tumors of the central nervous system are The NCCN Clinical Practice Guidelines in Oncology a heterogeneous group of neoplasms with varied outcomes and (NCCN Guidelines®) are a statement of consensus of the management strategies. -

Malignant Glioma Arising at the Site of an Excised Cerebellar Hemangioblastoma After Irradiation in a Von Hippel-Lindau Disease Patient
DOI 10.3349/ymj.2009.50.4.576 Case Report pISSN: 0513-5796, eISSN: 1976-2437 Yonsei Med J 50(4): 576-581, 2009 Malignant Glioma Arising at the Site of an Excised Cerebellar Hemangioblastoma after Irradiation in a von Hippel-Lindau Disease Patient Na-Hye Myong1 and Bong-Jin Park2 1Department of Pathology, Dankook University College of Medicine, Cheonan; 2Department of Neurosurgery, Kyunghee University Hospital, Seoul, Korea. We describe herein a malignant glioma arising at the site of the resected hemangioblastoma after irradiation in a patient with von Hippel-Lindau disease (VHL). The patient was a 25 year-old male with multiple heman- gioblastomas at the cerebellum and spinal cord, multiple pancreatic cysts and a renal cell carcinoma; he was diagnosed as having VHL disease. The largest hemangioblastoma at the right cerebellar hemisphere was completely removed, and he received high-dose irradiation postoperatively. The tumor recurred at the same site 7 years later, which was a malignant glioma with no evidence of hemangioblastoma. The malignant glioma showed molecular genetic profiles of radiation-induced tumors because of its diffuse p53 immunostaining and the loss of p16 immunoreactivity. The genetic study to find the loss of heterozygosity (LOH) of VHL gene revealed that only the cerebellar hemangioblastoma showed allelic losses for the gene. To the best of our knowledge, this report is the first to show a malignant glioma that developed in a patient with VHL disease after radiation therapy at the site of an excised hemangioblastoma. This report also suggests that radiation therapy should be performed very carefully in VHL patients with hemangioblastomas. -

Charts Chart 1: Benign and Borderline Intracranial and CNS Tumors Chart
Charts Chart 1: Benign and Borderline Intracranial and CNS Tumors Chart Glial Tumor Neuronal and Neuronal‐ Ependymomas glial Neoplasms Subependymoma Subependymal Giant (9383/1) Cell Astrocytoma(9384/1) Myyppxopapillar y Desmoplastic Infantile Ependymoma Astrocytoma (9412/1) (9394/1) Chart 1: Benign and Borderline Intracranial and CNS Tumors Chart Glial Tumor Neuronal and Neuronal‐ Ependymomas glial Neoplasms Subependymoma Subependymal Giant (9383/1) Cell Astrocytoma(9384/1) Myyppxopapillar y Desmoplastic Infantile Ependymoma Astrocytoma (9412/1) (9394/1) Use this chart to code histology. The tree is arranged Chart Instructions: Neuroepithelial in descending order. Each branch is a histology group, starting at the top (9503) with the least specific terms and descending into more specific terms. Ependymal Embryonal Pineal Choro id plexus Neuronal and mixed Neuroblastic Glial Oligodendroglial tumors tumors tumors tumors neuronal-glial tumors tumors tumors tumors Pineoblastoma Ependymoma, Choroid plexus Olfactory neuroblastoma Oligodendroglioma NOS (9391) (9362) carcinoma Ganglioglioma, anaplastic (9522) NOS (9450) Oligodendroglioma (9390) (9505 Olfactory neurocytoma Ganglioglioma, malignant (()9521) anaplastic (()9451) Anasplastic ependymoma (9505) Olfactory neuroepithlioma Oligodendroblastoma (9392) (9523) (9460) Papillary ependymoma (9393) Glioma, NOS (9380) Supratentorial primitive Atypical EdEpendymo bltblastoma MdllMedulloep ithliithelioma Medulloblastoma neuroectodermal tumor tetratoid/rhabdoid (9392) (9501) (9470) (PNET) (9473) tumor -

Paraganglioma (PGL) Tumors in Patients with Succinate Dehydrogenase-Related PCC–PGL Syndromes: a Clinicopathological and Molecular Analysis
T G Papathomas and others Non-PCC/PGL tumors in the SDH 170:1 1–12 Clinical Study deficiency Non-pheochromocytoma (PCC)/paraganglioma (PGL) tumors in patients with succinate dehydrogenase-related PCC–PGL syndromes: a clinicopathological and molecular analysis Thomas G Papathomas1, Jose Gaal1, Eleonora P M Corssmit2, Lindsey Oudijk1, Esther Korpershoek1, Ketil Heimdal3, Jean-Pierre Bayley4, Hans Morreau5, Marieke van Dooren6, Konstantinos Papaspyrou7, Thomas Schreiner8, Torsten Hansen9, Per Arne Andresen10, David F Restuccia1, Ingrid van Kessel6, Geert J L H van Leenders1, Johan M Kros1, Leendert H J Looijenga1, Leo J Hofland11, Wolf Mann7, Francien H van Nederveen12, Ozgur Mete13,14, Sylvia L Asa13,14, Ronald R de Krijger1,15 and Winand N M Dinjens1 1Department of Pathology, Josephine Nefkens Institute, Erasmus MC, University Medical Center, PO Box 2040, 3000 CA Rotterdam, The Netherlands, 2Department of Endocrinology, Leiden University Medical Center, Leiden,The Netherlands, 3Section for Clinical Genetics, Department of Medical Genetics, Oslo University Hospital, Oslo, Norway, 4Department of Human and Clinical Genetics, Leiden University Medical Center, Leiden, The Netherlands, 5Department of Pathology, Leiden University Medical Center, Leiden, The Netherlands, 6Department of Clinical Genetics, Erasmus MC, University Medical Center, Rotterdam, The Netherlands, 7Department of Otorhinolaryngology, Head and Neck Surgery, University Medical Center of the Johannes Gutenberg University Mainz, Mainz, Germany, 8Section for Specialized Endocrinology, -

Central Nervous System Tumors General ~1% of Tumors in Adults, but ~25% of Malignancies in Children (Only 2Nd to Leukemia)
Last updated: 3/4/2021 Prepared by Kurt Schaberg Central Nervous System Tumors General ~1% of tumors in adults, but ~25% of malignancies in children (only 2nd to leukemia). Significant increase in incidence in primary brain tumors in elderly. Metastases to the brain far outnumber primary CNS tumors→ multiple cerebral tumors. One can develop a very good DDX by just location, age, and imaging. Differential Diagnosis by clinical information: Location Pediatric/Young Adult Older Adult Cerebral/ Ganglioglioma, DNET, PXA, Glioblastoma Multiforme (GBM) Supratentorial Ependymoma, AT/RT Infiltrating Astrocytoma (grades II-III), CNS Embryonal Neoplasms Oligodendroglioma, Metastases, Lymphoma, Infection Cerebellar/ PA, Medulloblastoma, Ependymoma, Metastases, Hemangioblastoma, Infratentorial/ Choroid plexus papilloma, AT/RT Choroid plexus papilloma, Subependymoma Fourth ventricle Brainstem PA, DMG Astrocytoma, Glioblastoma, DMG, Metastases Spinal cord Ependymoma, PA, DMG, MPE, Drop Ependymoma, Astrocytoma, DMG, MPE (filum), (intramedullary) metastases Paraganglioma (filum), Spinal cord Meningioma, Schwannoma, Schwannoma, Meningioma, (extramedullary) Metastases, Melanocytoma/melanoma Melanocytoma/melanoma, MPNST Spinal cord Bone tumor, Meningioma, Abscess, Herniated disk, Lymphoma, Abscess, (extradural) Vascular malformation, Metastases, Extra-axial/Dural/ Leukemia/lymphoma, Ewing Sarcoma, Meningioma, SFT, Metastases, Lymphoma, Leptomeningeal Rhabdomyosarcoma, Disseminated medulloblastoma, DLGNT, Sellar/infundibular Pituitary adenoma, Pituitary adenoma, -

A Rare Disease in Pediatric Patients
Cartas ao Editor Jornal de Pediatria - Vol. 85, Nº 3, 2009 277 Resposta do autor Gliomatose leptomeníngea primária difusa: uma doença rara em pacientes pediátricos Prezado Editor, Recebemos com prazer os comentários acima. Os autores esclarecem que, em momento algum, focaram suas preo- Prezado Editor, cupações em critérios para definir síndrome metabólica, até mesmo porque se ela existe1, não existem critérios de consenso O recente relato de caso de Val Filho & Avelar1 sobre um para o diagnóstico em adultos2,3 tampouco em adolescentes. raro tumor cerebral pediátrico é interessante e merece grande Preocupamo-nos sim, em mostrar a presença de fatores de consideração no contexto da literatura publicada sobre esse risco em uma amostra selecionada ao acaso, que eles existem assunto, já que representa um evento muito incomum. Em em proporções tais que podem até ser agrupados, e, por um nome da precisão científica, devemos, portanto, fazer uma dos muitos critérios sugeridos na literatura, com valores de importante correção no que se refere ao diagnóstico. referências pediátricos, dar origem ao que se denomina sín- Os autores fizeram uma revisão breve e adequada sobre a drome metabólica. Não nos propusemos, portanto, a estudar gliomatose cerebral; uma doença rara, classificada pela edição associações entre fatores de risco. de 2007 da Organização Mundial da Saúde (OMS)2 como uma Os autores acreditam que discutir critérios para o que não lesão de grau III com prognóstico reservado. A gliomatose 2,3 existe consenso é desfocar a atenção da gravidade -

6 Magnetic Resonance Imaging
Chapter 6 / MRI 105 6 Magnetic Resonance Imaging Paul M. Ruggieri Summary Magnetic resonance (MR) is clearly the accepted imaging standard for the preliminary evalua- tion, peri-operative management, and routine longitudinal follow-up of patients with high-grade glio- mas (HGG). The purpose of this chapter is to review the imaging characteristics of HGG using conventional MR imaging techniques. Whereas the newer techniques of MR diffusion, perfusion, diffusion tensor imaging, and MR spectroscopy will be included as part of this discussion of the high grade neoplasms, the detailed concepts of such studies will be discussed elsewhere in this text. Key Words: MR imaging; anaplastic astrocytoma; glioblastoma multiforme; gliosarcoma; gliomatosis cerebri; oligodendroglioma. INTRODUCTION Magnetic resonance (MR) is clearly the accepted imaging standard for the preliminary evaluation, peri-operative management, and routine longitudinal follow-up of patients with high-grade gliomas (HGG). The purpose of this chapter is to review the imaging characteristics of HGG using conventional MR imaging techniques. Whereas the newer techniques of MR diffusion, perfusion, diffusion tensor imaging, and MR spectroscopy will be included as part of this discussion of the high grade neoplasms, the detailed concepts of such studies will be discussed elsewhere in this text. In general terms, high-grade glial neoplasms are conventionally thought of as infiltrative parenchymal masses that are hyperintense on FLAIR fluid-attenuated inversion recovery (FLAIR) and T2-weighted images, hypointense on unenhanced T1-weighted images, may or may not extend into the corpus callosum, are surrounded by extensive vasogenic edema, and prominently enhance following gadolinium administration. It must be pointed out that this description is clearly just a generalization as many high-grade neoplasms clearly do not follow these “rules” and some of these characteristics may even be seen in low-grade neoplasms. -

Ambient Mass Spectrometry for the Intraoperative Molecular Diagnosis of Human Brain Tumors
Ambient mass spectrometry for the intraoperative molecular diagnosis of human brain tumors Livia S. Eberlina, Isaiah Nortonb, Daniel Orringerb, Ian F. Dunnb, Xiaohui Liub, Jennifer L. Ideb, Alan K. Jarmuscha, Keith L. Ligonc, Ferenc A. Joleszd, Alexandra J. Golbyb,d, Sandro Santagatac, Nathalie Y. R. Agarb,d,1, and R. Graham Cooksa,1 aDepartment of Chemistry and Center for Analytical Instrumentation Development, Purdue University, West Lafayette, IN 47907; and Departments of bNeurosurgery, cPathology, and dRadiology, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02115 Edited by Jack Halpern, The University of Chicago, Chicago, IL, and approved December 5, 2012 (received for review September 11, 2012) The main goal of brain tumor surgery is to maximize tumor resection at Brigham and Women’s Hospital (BWH), created an opportu- while preserving brain function. However, existing imaging and nity for collecting information about the extent of tumor resection surgical techniques do not offer the molecular information needed during surgery (5, 6). Although brain tumor resection typically to delineate tumor boundaries. We have developed a system to requires multiple hours, intraoperative MRI can be completed rapidly analyze and classify brain tumors based on lipid information and information evaluated within an hour. However, MRI has acquired by desorption electrospray ionization mass spectrometry limited ability to distinguish residual tumor from surrounding (DESI-MS). In this study, a classifier was built to discriminate gliomas normal brain (9). In consequence, there is a need for more de- and meningiomas based on 36 glioma and 19 meningioma samples. tailed molecular information to be acquired on a timescale closer The classifier was tested and results were validated for intraoper- to real time than can be supplied by MRI. -
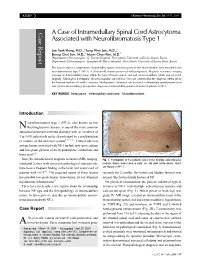
A Case of Intramedullary Spinal Cord Astrocytoma Associated with Neurofibromatosis Type 1
KISEP J Korean Neurosurg Soc 36 : 69-71, 2004 Case Report A Case of Intramedullary Spinal Cord Astrocytoma Associated with Neurofibromatosis Type 1 Jae Taek Hong, M.D.,1 Sang Won Lee, M.D.,1 Byung Chul Son, M.D.,1 Moon Chan Kim, M.D.2 Department of Neurosurgery,1 St. Vincent Hospital, The Catholic University of Korea, Suwon, Korea Department of Neurosurgery,2 Kangnam St. Mary's Hospital, The Catholic University of Korea, Seoul, Korea The authors report a symptomatic intramedullary spinal cord astrocytoma in the thoracolumbar area associated with neurofibromatosis type 1 (NF-1). A 38-year-old woman presented with paraparesis. Magnetic resonance imaging revealed an intramedullary lesion within the lower thoracic spinal cord and conus medullaris, which was removed surgically. Pathological investigation showed anaplastic astrocytoma. This case confirms that the diagnosis criteria set by the National Institute of Health Consensus Development Conference can be useful to differentiate ependymoma from astrocytoma when making a preoperative diagnosis of intramedullary spinal cord tumor in patients of NF-1. KEY WORDS : Astrocytoma·Intramedullary cord tumor·Neurofibromatosis. Introduction eurofibromatosis type 1 (NF-1), also known as von N Recklinghausen's disease, is one of the most common autosomal dominant inherited disorders with an incidence of 1 in 3,000 individuals and is characterized by a predisposition to tumors of the nervous system5,6,12,16). Central nervous system lesions associated with NF-1 include optic nerve glioma and low-grade gliomas of the hypothalamus, cerebellum and brain stem6,10). Since the introduction of magnetic resonance(MR) imaging, Fig. 1. Photograph of the patient's back shows multiple subcutaneous incidental lesions with uncertain pathological characteristic nodules (black arrow) and a cafe-au-lait spot (white arrow), which have been a frequent finding in the brain and spinal cord of are typical of NF-1. -

Central Neurocytoma SNP Array Analyses, Subtel FISH, and Review
Pathology - Research and Practice 215 (2019) 152397 Contents lists available at ScienceDirect Pathology - Research and Practice journal homepage: www.elsevier.com/locate/prp Case report Central neurocytoma: SNP array analyses, subtel FISH, and review of the T literature Caroline Sandera,1, Marco Wallenborna,b,1, Vivian Pascal Brandtb, Peter Ahnertc, Vera Reuscheld, ⁎ Christan Eisenlöffele, Wolfgang Kruppa, Jürgen Meixensbergera, Heidrun Hollandb, a Dept. of Neurosurgery, University of Leipzig, Liebigstraße 26, 04103 Leipzig, Germany b Saxonian Incubator for Clinical Translation, University of Leipzig, Philipp-Rosenthal Str. 55, 04103 Leipzig, Germany c Institute for Medical Informatics, Statistics and Epidemiology, University of Leipzig, Haertelstraße 16-18, 04107 Leipzig, Germany d Dept. of Neuroradiology, University of Leipzig, Liebigstraße 22a, 04103 Leipzig, Germany e Dept. of Neuropathology, University of Leipzig, Liebigstraße 26, 04103 Leipzig, Germany ARTICLE INFO ABSTRACT Keywords: The central neurocytoma (CN) is a rare brain tumor with a frequency of 0.1-0.5% of all brain tumors. According Central neurocytoma to the World Health Organization classification, it is a benign grade II tumor with good prognosis. However, Cytogenetics some CN occur as histologically “atypical” variant, combined with increasing proliferation and poor clinical SNP array outcome. Detailed genetic knowledge could be helpful to characterize a potential atypical behavior in CN. Only FISH few publications on genetics of CN exist in the literature. Therefore, we performed cytogenetic analysis of an intraventricular neurocytoma WHO grade II in a 39-year-old male patient by use of genome-wide high-density single nucleotide polymorphism array (SNP array) and subtelomere FISH. Applying these techniques, we could detect known chromosomal aberrations and identified six not previously described chromosomal aberrations, gains of 1p36.33-p36.31, 2q37.1-q37.3, 6q27, 12p13.33-p13.31, 20q13.31-q13.33, and loss of 19p13.3-p12. -

A Case of Mushroom‑Shaped Anaplastic Oligodendroglioma Resembling Meningioma and Arteriovenous Malformation: Inadequacies of Diagnostic Imaging
EXPERIMENTAL AND THERAPEUTIC MEDICINE 10: 1499-1502, 2015 A case of mushroom‑shaped anaplastic oligodendroglioma resembling meningioma and arteriovenous malformation: Inadequacies of diagnostic imaging YAOLING LIU1,2, KANG YANG1, XU SUN1, XINYU LI1, MINGHAI WEI1, XIANG'EN SHI2, NINGWEI CHE1 and JIAN YIN1 1Department of Neurosurgery, The Second Affiliated Hospital of Dalian Medical University, Dalian, Liaoning 116044; 2Department of Neurosurgery, Affiliated Fuxing Hospital, The Capital University of Medical Sciences, Beijing 100038, P.R. China Received December 29, 2014; Accepted June 29, 2015 DOI: 10.3892/etm.2015.2676 Abstract. Magnetic resonance imaging (MRI) is the most tomas (WHO IV) (2). The median survival times of patients widely discussed and clinically employed method for the with WHO II and WHO III oligodendrogliomas are 9.8 and differential diagnosis of oligodendrogliomas; however, 3.9 years, respectively (1,2), and 6.3 and 2.8 years, respec- MRI occasionally produces unclear results that can hinder tively, if mixed with astrocytes (3,4). Surgical excision and a definitive oligodendroglioma diagnosis. The present study postoperative adjuvant radiotherapy is the traditional therapy describes the case of a 34-year-old man that suffered from for oligodendroglioma; however, studies have observed that, headache and right upper‑extremity weakness for 2 months. among intracranial tumors, anaplastic oligodendrogliomas Based on the presurgical evaluation, it was suggested that the are particularly sensitive to chemotherapy, and the prognosis patient had a World Health Organization (WHO) grade II-II of patients treated with chemotherapy is more favorable glioma, meningioma or arteriovenous malformation (AVM), than that of patients treated with radiotherapy (5‑7). -

Risk-Adapted Therapy for Young Children with Embryonal Brain Tumors, High-Grade Glioma, Choroid Plexus Carcinoma Or Ependymoma (Sjyc07)
SJCRH SJYC07 CTG# - NCT00602667 Initial version, dated: 7/25/2007, Resubmitted to CPSRMC 9/24/2007 and 10/6/2007 (IRB Approved: 11/09/2007) Activation Date: 11/27/2007 Amendment 1.0 dated January 23, 2008, submitted to CPSRMC: January 23, 2008, IRB Approval: March 10, 2008 Amendment 2.0 dated April 16, 2008, submitted to CPSRMC: April 16, 2008, (IRB Approval: May 13, 2008) Revision 2.1 dated April 29, 2009 (IRB Approved: April 30, 2009 ) Amendment 3.0 dated June 22, 2009, submitted to CPSRMC: June 22, 2009 (IRB Approved: July 14, 2009) Activated: August 11, 2009 Amendment 4.0 dated March 01, 2010 (IRB Approved: April 20, 2010) Activated: May 3, 2010 Amendment 5.0 dated July 19, 2010 (IRB Approved: Sept 17, 2010) Activated: September 24, 2010 Amendment 6.0 dated August 27, 2012 (IRB approved: September 24, 2012) Activated: October 18, 2012 Amendment 7.0 dated February 22, 2013 (IRB approved: March 13, 2013) Activated: April 4, 2013 Amendment 8.0 dated March 20, 2014. Resubmitted to IRB May 20, 2014 (IRB approved: May 22, 2014) Activated: May 30, 2014 Amendment 9.0 dated August 26, 2014. (IRB approved: October 14, 2014) Activated: November 4, 2014 Un-numbered revision dated March 22, 2018. (IRB approved: March 27, 2018) Un-numbered revision dated October 22, 2018 (IRB approved: 10-24-2018) RISK-ADAPTED THERAPY FOR YOUNG CHILDREN WITH EMBRYONAL BRAIN TUMORS, HIGH-GRADE GLIOMA, CHOROID PLEXUS CARCINOMA OR EPENDYMOMA (SJYC07) Principal Investigator Amar Gajjar, M.D. Division of Neuro-Oncology Department of Oncology Section Coordinators David Ellison, M.D., Ph.D.