Incorporation Into the Prereplicative Complex Activates the Mcm2–7 Helicase for Cdc7–Dbf4 Phosphorylation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Regulation of Calcium/Calmodulin-Dependent Protein Kinase II Activation by Intramolecular and Intermolecular Interactions
8394 • The Journal of Neuroscience, September 29, 2004 • 24(39):8394–8398 Mini-Review Regulation of Calcium/Calmodulin-Dependent Protein Kinase II Activation by Intramolecular and Intermolecular Interactions Leslie C. Griffith Department of Biology and Volen Center for Complex Systems, Brandeis University, Waltham, Massachusetts 02454-9110 Key words: calcium; calmodulin; learning; localization; NMDA; phosphatase; protein kinase As its name implies, calcium/calmodulin-dependent protein ki- The overlap of these subdomains is no accident. Binding of nase II (CaMKII) is calcium dependent. In its basal state, the Ca 2ϩ/CaM is the primary signal for release of autoinhibition. activity of CaMKII is extremely low. Regulation of intracellular Current models of activation posit that the binding of Ca 2ϩ/CaM calcium levels allows the neuron to link activity with phosphor- serves to disrupt the interactions of specific residues within the ylation by CaMKII. This review will briefly summarize our cur- autoinhibitory domain with the catalytic domain (Smith et al., rent understanding of the intramolecular mechanisms of activity 1992). Because there is no crystal structure for the catalytic and regulation and their modulation by Ca 2ϩ/CaM and will then regulatory parts of CaMKII, the interaction face of these two focus on the growing number of other modes of intermolecular domains has been inferred using the effects of charge-reversal regulation of CaMKII activity by substrate and scaffolding mutagenesis on activity and molecular modeling (Yang and molecules. Schulman, 1999). This study confirmed the role of Arg 297 at the P-3 position of the pseudosubstrate ligand (Mukherji and Soder- Regulation of CaMKII by its autoinhibitory domain ling, 1995) and identified residues in the catalytic domain that All members of the CaMKII family (␣, , ␥, and ␦ isozymes) share may have direct interactions with the regulatory region. -

Mcm2 Is a Target of Regulation by Cdc7–Dbf4 During the Initiation of DNA Synthesis
Downloaded from genesdev.cshlp.org on September 25, 2021 - Published by Cold Spring Harbor Laboratory Press Mcm2 is a target of regulation by Cdc7–Dbf4 during the initiation of DNA synthesis Ming Lei,1 Yasuo Kawasaki,1,2 Michael R. Young,1,3 Makoto Kihara,2 Akio Sugino,2 and Bik K. Tye1,4 1Section of Biochemistry, Molecular and Cell Biology, Cornell University, Ithaca, New York 14853-2703 USA; 2Research Institute for Microbial Diseases, Osaka University, Osaka 565, Japan The initiation of DNA synthesis is an important cell cycle event that defines the beginning of S phase. This critical event involves the participation of proteins whose functions are regulated by cyclin dependent protein kinases (Cdks). The Mcm2–7 proteins are a family of six conserved proteins that are essential for the initiation of DNA synthesis in all eukaryotes. In Saccharomyces cerevisiae, members of the Mcm2–7 family undergo cell cycle-specific phosphorylation. Phosphorylation of Mcm proteins at the beginning of S phase coincides with the removal of these proteins from chromatin and the onset of DNA synthesis. In this study, we identified DBF4, which encodes the regulatory subunit of a Cdk-like protein kinase Cdc7–Dbf4, in a screen for second site suppressors of mcm2-1. The dbf4 suppressor mutation restores competence to initiate DNA synthesis to the mcm2-1 mutant. Cdc7–Dbf4 interacts physically with Mcm2 and phosphorylates Mcm2 and three other members of the Mcm2–7 family in vitro. Blocking the kinase activity of Cdc7–Dbf4 at the G1-to-S phase transition also blocks the phosphorylation of Mcm2 at this defined point of the cell cycle. -

Saccharomyces Rrm3p, a 5 to 3 DNA Helicase That Promotes Replication
Downloaded from genesdev.cshlp.org on September 24, 2021 - Published by Cold Spring Harbor Laboratory Press Saccharomyces Rrm3p, a 5 to 3 DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA Andreas S. Ivessa,1 Jin-Qiu Zhou,1,2 Vince P. Schulz, Ellen K. Monson, and Virginia A. Zakian3 Department of Molecular Biology, Princeton University, Princeton, New Jersey 08544-1014, USA In wild-type Saccharomyces cerevisiae, replication forks slowed during their passage through telomeric C1–3A/TG1–3 tracts. This slowing was greatly exacerbated in the absence of RRM3, shown here to encode a 5 ,to 3 DNA helicase. Rrm3p-dependent fork progression was seen at a modified Chromosome VII-L telomere at the natural X-bearing Chromosome III-L telomere, and at Y-bearing telomeres. Loss of Rrm3p also resulted in replication fork pausing at specific sites in subtelomeric DNA, such as at inactive replication origins, and at internal tracts of C1–3A/TG1–3 DNA. The ATPase/helicase activity of Rrm3p was required for its role in telomeric and subtelomeric DNA replication. Because Rrm3p was telomere-associated in vivo, it likely has a direct role in telomere replication. [Key Words: Telomere; helicase; telomerase; replication; RRM3; yeast] Received February 7, 2002; revised version accepted April 10, 2002. Telomeres are the natural ends of eukaryotic chromo- Because conventional DNA polymerases cannot repli- somes. In most organisms, the very ends of chromo- cate the very ends of linear DNA molecules, special somes consist of simple repeated sequences. For ex- mechanisms are required to prevent the loss of terminal ample, Saccharomyces cerevisiae chromosomes end in DNA. -
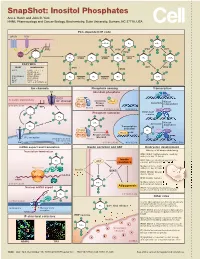
Snapshot: Inositol Phosphates Ace J
SnapShot: Inositol Phosphates Ace J. Hatch and John D. York HHMI, Pharmacology and Cancer Biology, Biochemistry, Duke University, Durham, NC 27710, USA PLC-dependent IP code GPCR RTK O O O O O O 5-PP-IP4 IP4 5-IP7 O O O O O O PIP2 O IP6K O IP6K O VIP1 O O O 2 O ITPK1 O 13 O PLC 2 O O O O O O O O 4 6 13 O 5 IP3 IPMK IP4 IPMK IP5 IPK1 IP6 1,5-IP8 4 6 O O 5 O O O O O O O O ENZYMES O O O O O O YEAST MAMMALIAN IP3K VIP1 IP6K IPMK PLC1 PLCβ, γ, δ, ε, ζ, η O - IP3KA, B, C - ITPK1 (IP56K) O O O O O O O O IPK2(ARG82) IPMK (IPK2) IP4 IP3 IP4 1-IP7 IPK1 IPK1 (IP5K) INPP5 ITPK1 KCS1 IP6K1, 2, 3 O O O O O O VIP1 VIP1, 2 (PPIP5K1, 2) O O Ion channels Phosphate sensing Transcription Cl- Abundant phosphate MCM1 ARG80 CIC3 P PLASMA MEMBRANE - Pho80 Cl channel Pho4 Kinase Kinase Assembly Pho85 independent CYTOPLASM activity 2 O PIP2 Pho81 13 CYTOPLASM NUCLEUS IPK2 ARG81 4 6 Phosphate starvation MCM1-ArgR O 5 O complex O O IP4 O O O O O O O 1-IP7 Kinase Activation dependent IP3 O O Transcription O O O activated Pho80 IP4 O X Pho4 O O Pho85 Kinase activity IP receptor blocked O 3 ENDOPLASMIC Pho81 RETICULUM Ca2+ CYTOPLASM NUCLEUS NUCLEUS mRNA export and translation Insulin secretion and AKT Embryonic development Translation termination Effects of IP kinase deficiency O IPMK (IPK2): Multiple defects, death by embryonic day 10 (mice) O O Insulin IPK1: Cillia are shortened and immotile IP6 AKT resistance causing patterning defects (zebrash) O O Multiple defects, death by Ribosome O embryonic day 8.5 (mice) GleI eRF1 Insulin GSK3β Dbp5 ITPK1 (IP56K): Neural tube -

The Architecture of a Eukaryotic Replisome
The Architecture of a Eukaryotic Replisome Jingchuan Sun1,2, Yi Shi3, Roxana E. Georgescu3,4, Zuanning Yuan1,2, Brian T. Chait3, Huilin Li*1,2, Michael E. O’Donnell*3,4 1 Biosciences Department, Brookhaven National Laboratory, Upton, New York, USA 2 Department of Biochemistry & Cell Biology, Stony Brook University, Stony Brook, New York, USA. 3 The Rockefeller University, 1230 York Avenue, New York, New York, USA. 4 Howard Hughes Medical Institute *Correspondence and requests for materials should be addressed to M.O.D. ([email protected]) or H.L. ([email protected]) ABSTRACT At the eukaryotic DNA replication fork, it is widely believed that the Cdc45-Mcm2-7-GINS (CMG) helicase leads the way in front to unwind DNA, and that DNA polymerases (Pol) trail behind the helicase. Here we use single particle electron microscopy to directly image a replisome. Contrary to expectations, the leading strand Pol ε is positioned ahead of CMG helicase, while Ctf4 and the lagging strand Pol α-primase (Pol α) are behind the helicase. This unexpected architecture indicates that the leading strand DNA travels a long distance before reaching Pol ε, it first threads through the Mcm2-7 ring, then makes a U-turn at the bottom to reach Pol ε at the top of CMG. Our work reveals an unexpected configuration of the eukaryotic replisome, suggests possible reasons for this architecture, and provides a basis for further structural and biochemical replisome studies. INTRODUCTION DNA is replicated by a multi-protein machinery referred to as a replisome 1,2. Replisomes contain a helicase to unwind DNA, DNA polymerases that synthesize the leading and lagging strands, and a primase that makes short primed sites to initiate DNA synthesis on both strands. -

Three Dact Gene Family Members Are Expressed During Embryonic Development
Three Dact Gene Family Members are Expressed During Embryonic Development and in the Adult Brains of Mice Daniel A Fisher, Saul Kivimäe, Jun Hoshino, Rowena Suriben, Pierre -Marie Martin, Nichol Baxter, Benjamin NR Cheyette Department of Psychiatry & Gra duate Programs in Developmental Biology and Neuroscience, University of California, San Francisco, 94143-2611 Correspondence: Benjamin NR Cheyette [email protected] 415.476.7826 Running Title: Mouse Dact Gene Family Expression Key Words: mouse, Dpr, Frodo, Thyex, Dact, Wnt, Dvl, expression, embryo, brain Supported by: NIH: MH01750 K08; NARSAD Young Investigator Award; NAAR award #551. Abstract Members of the Dact protein family were initially identified through binding to Dishevelled (Dvl), a cytoplasmic protein central to Wnt signaling. During mouse development, Dact1 is detected in the presomitic mesoderm and somites during segmentation, in the limb bud mesenchyme and other mesoderm-derived tissues, and in the central nervous system (CNS). Dact2 expression is most prominent during organogenesis of the thymus, kidneys, and salivary glands, with much lower levels in the somites and in the developing CNS. Dact3, not previously described in any organism, is expressed in the ventral region of maturing somites, limb bud and branchial arch mesenchyme, and in the embryonic CNS; of the three paralogs it is the most highly expressed in the adult cerebral cortex. These data are consistent with studies in other vertebrates showing that Dact paralogs have distinct signaling and developmental roles, and suggest they may differentially contribute to postnatal brain physiology. Introduction Signaling downstream of secreted Wnt ligands is a conserved process in multicellular animals that plays important roles during development and, when misregulated, contributes to cancer and other diseases (Polakis, 2000; Moon et al., 2002). -

DNA Replication Stress Response Involving PLK1, CDC6, POLQ
DNA replication stress response involving PLK1, CDC6, POLQ, RAD51 and CLASPIN upregulation prognoses the outcome of early/mid-stage non-small cell lung cancer patients C. Allera-Moreau, I. Rouquette, B. Lepage, N. Oumouhou, M. Walschaerts, E. Leconte, V. Schilling, K. Gordien, L. Brouchet, Mb Delisle, et al. To cite this version: C. Allera-Moreau, I. Rouquette, B. Lepage, N. Oumouhou, M. Walschaerts, et al.. DNA replica- tion stress response involving PLK1, CDC6, POLQ, RAD51 and CLASPIN upregulation prognoses the outcome of early/mid-stage non-small cell lung cancer patients. Oncogenesis, Nature Publishing Group: Open Access Journals - Option C, 2012, 1, pp.e30. 10.1038/oncsis.2012.29. hal-00817701 HAL Id: hal-00817701 https://hal.archives-ouvertes.fr/hal-00817701 Submitted on 9 Jun 2021 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution - NonCommercial - NoDerivatives| 4.0 International License Citation: Oncogenesis (2012) 1, e30; doi:10.1038/oncsis.2012.29 & 2012 Macmillan Publishers Limited All rights reserved 2157-9024/12 www.nature.com/oncsis ORIGINAL ARTICLE DNA replication stress response involving PLK1, CDC6, POLQ, RAD51 and CLASPIN upregulation prognoses the outcome of early/mid-stage non-small cell lung cancer patients C Allera-Moreau1,2,7, I Rouquette2,7, B Lepage3, N Oumouhou3, M Walschaerts4, E Leconte5, V Schilling1, K Gordien2, L Brouchet2, MB Delisle1,2, J Mazieres1,2, JS Hoffmann1, P Pasero6 and C Cazaux1 Lung cancer is the leading cause of cancer deaths worldwide. -

Regulation of Calmodulin-Stimulated Cyclic Nucleotide Phosphodiesterase (PDE1): Review
95-105 5/6/06 13:44 Page 95 INTERNATIONAL JOURNAL OF MOLECULAR MEDICINE 18: 95-105, 2006 95 Regulation of calmodulin-stimulated cyclic nucleotide phosphodiesterase (PDE1): Review RAJENDRA K. SHARMA, SHANKAR B. DAS, ASHAKUMARY LAKSHMIKUTTYAMMA, PONNIAH SELVAKUMAR and ANURAAG SHRIVASTAV Department of Pathology and Laboratory Medicine, College of Medicine, University of Saskatchewan, Cancer Research Division, Saskatchewan Cancer Agency, 20 Campus Drive, Saskatoon SK S7N 4H4, Canada Received January 16, 2006; Accepted March 13, 2006 Abstract. The response of living cells to change in cell 6. Differential inhibition of PDE1 isozymes and its environment depends on the action of second messenger therapeutic applications molecules. The two second messenger molecules cAMP and 7. Role of proteolysis in regulating PDE1A2 Ca2+ regulate a large number of eukaryotic cellular events. 8. Role of PDE1A1 in ischemic-reperfused heart Calmodulin-stimulated cyclic nucleotide phosphodiesterase 9. Conclusion (PDE1) is one of the key enzymes involved in the complex interaction between cAMP and Ca2+ second messenger systems. Some PDE1 isozymes have similar kinetic and 1. Introduction immunological properties but are differentially regulated by Ca2+ and calmodulin. Accumulating evidence suggests that the A variety of cellular activities are regulated through mech- activity of PDE1 is selectively regulated by cross-talk between anisms controlling the level of cyclic nucleotides. These Ca2+ and cAMP signalling pathways. These isozymes are mechanisms include synthesis, degradation, efflux and seque- also further distinguished by various pharmacological agents. stration of cyclic adenosine 3':5'-monophosphate (cAMP) and We have demonstrated a potentially novel regulation of PDE1 cyclic guanosine 3':5'- monophosphate (cGMP) within the by calpain. -

The Biochemical Activities of the Saccharomyces Cerevisiae Pif1 Helicase Are Regulated by Its N-Terminal Domain
G C A T T A C G G C A T genes Article The Biochemical Activities of the Saccharomyces cerevisiae Pif1 Helicase Are Regulated by Its N-Terminal Domain David G. Nickens y, Christopher W. Sausen y and Matthew L. Bochman * Molecular and Cellular Biochemistry Department, Indiana University, Bloomington, IN 47405, USA; [email protected] (D.G.N.); [email protected] (C.W.S.) * Correspondence: [email protected] These authors contributed equally to this work. y Received: 31 March 2019; Accepted: 20 May 2019; Published: 28 May 2019 Abstract: Pif1 family helicases represent a highly conserved class of enzymes involved in multiple aspects of genome maintenance. Many Pif1 helicases are multi-domain proteins, but the functions of their non-helicase domains are poorly understood. Here, we characterized how the N-terminal domain (NTD) of the Saccharomyces cerevisiae Pif1 helicase affects its functions both in vivo and in vitro. Removal of the Pif1 NTD alleviated the toxicity associated with Pif1 overexpression in yeast. Biochemically, the N-terminally truncated Pif1 (Pif1DN) retained in vitro DNA binding, DNA unwinding, and telomerase regulation activities, but these activities differed markedly from those displayed by full-length recombinant Pif1. However, Pif1DN was still able to synergize with the Hrq1 helicase to inhibit telomerase activity in vitro, similar to full-length Pif1. These data impact our understanding of Pif1 helicase evolution and the roles of these enzymes in the maintenance of genome integrity. Keywords: DNA helicase; Saccharomyces cerevisiae; Pif1; telomerase; telomere 1. Introduction DNA helicases are enzymes that couple DNA binding and ATP hydrolysis to unwind double-stranded DNA (dsDNA) into its component single strands [1]. -

Protein Kinases Phosphorylation/Dephosphorylation Protein Phosphorylation Is One of the Most Important Mechanisms of Cellular Re
Protein Kinases Phosphorylation/dephosphorylation Protein phosphorylation is one of the most important mechanisms of cellular responses to growth, stress metabolic and hormonal environmental changes. Most mammalian protein kinases have highly a homologous 30 to 32 kDa catalytic domain. • Most common method of reversible modification - activation and localization • Up to 1/3 of cellular proteins can be phosphorylated • Leads to a very fast response to cellular stress, hormonal changes, learning processes, transcription regulation .... • Different than allosteric or Michealis Menten regulation Protein Kinome To date – 518 human kinases known • 50 kinase families between yeast, invertebrate and mammaliane kinomes • 518 human PKs, most (478) belong to single super family whose catalytic domain are homologous. • Kinase dendrogram displays relative similarities based on catalytic domains. • AGC (PKA, PKG, PKC) • CAMK (Casein kinase 1) • CMGC (CDC, MAPK, GSK3, CLK) • STE (Sterile 7, 11 & 20 kinases) • TK (Tryosine kinases memb and cyto) • TKL (Tyrosine kinase-like) • Phosphorylation stabilized thermodynamically - only half available energy used in adding phosphoryl to protein - change in free energy forces phosphorylation reaction in one direction • Phosphatases reverse direction • The rate of reaction of most phosphatases are 1000 times faster • Phosphorylation occurs on Ser/The or Tyr • What differences occur due to the addition of a phosphoryl group? • Regulation of protein phosphorylation varies depending on protein - some turned on or off -

Mcm10 Has Potent Strand-Annealing Activity and Limits Translocase-Mediated Fork Regression
Mcm10 has potent strand-annealing activity and limits translocase-mediated fork regression Ryan Maylea, Lance Langstona,b, Kelly R. Molloyc, Dan Zhanga, Brian T. Chaitc,1,2, and Michael E. O’Donnella,b,1,2 aLaboratory of DNA Replication, The Rockefeller University, New York, NY 10065; bHoward Hughes Medical Institute, The Rockefeller University, New York, NY 10065; and cLaboratory of Mass Spectrometry and Gaseous Ion Chemistry, The Rockefeller University, New York, NY 10065 Contributed by Michael E. O’Donnell, November 19, 2018 (sent for review November 8, 2018; reviewed by Zvi Kelman and R. Stephen Lloyd) The 11-subunit eukaryotic replicative helicase CMG (Cdc45, Mcm2-7, of function using genetics, cell biology, and cell extracts have GINS) tightly binds Mcm10, an essential replication protein in all identified Mcm10 functions in replisome stability, fork progres- eukaryotes. Here we show that Mcm10 has a potent strand- sion, and DNA repair (21–25). Despite significant advances in the annealing activity both alone and in complex with CMG. CMG- understanding of Mcm10’s functions, mechanistic in vitro studies Mcm10 unwinds and then reanneals single strands soon after they of Mcm10 in replisome and repair reactions are lacking. have been unwound in vitro. Given the DNA damage and replisome The present study demonstrates that Mcm10 on its own rap- instability associated with loss of Mcm10 function, we examined the idly anneals cDNA strands even in the presence of the single- effect of Mcm10 on fork regression. Fork regression requires the strand (ss) DNA-binding protein RPA, a property previously unwinding and pairing of newly synthesized strands, performed by associated with the recombination protein Rad52 (26). -

Huh7 HK4+ HK2- Cells a Protein Complementation Assay B Coimmunoprecipitation Even If NS3 Is Able to Stimulates Glycolysis in Cells Expressing Replicate
Dengue virus protein NS3 activates hexokinase activity in SAT-390 hepatocytes to support virus replication Marianne FIGL, Clémence JACQUEMIN, Patrice ANDRE, Laure PERRIN-COCON, Vincent LOTTEAU, Olivier DIAZ International Center for Infectiology Research (CIRI), INSERM U1111, CNRS UMR5308, Université de Lyon, FRANCE 1 INTRODUCTION 4 RESULTS 5 CONCLUSIONS Result 2: DENV NS3 protein interacts with hexokinases Viruses are mandatory parasites that use metabolism machinery to Result 1: DENV efficiently replicates in HuH7 HK4+ HK2- cells A Protein Complementation Assay B Coimmunoprecipitation Even if NS3 is able to stimulates glycolysis in cells expressing replicate. Growing literature demonstrates that viruses manipulate A.A. DENV-NS3 versus human metabolism enzymes DENVB.B.-NS3 versus hexokinases A. HuH7 HuH7 HK4+ HK2- B. (a) (b) 60 Lysate Co-IP HK2 or HK4, we observe an higher DENV replication in HuH7 central carbon metabolism (CCM) and more specifically glycolysis for HuH7 HuH7 55 NS3-3xFlag - + - + HuH7 HK4+ HK2- HK4+ HK2- suggesting that HK4 positive cells are more susceptible their propagation [1]. However, the underlying mechanisms are not HuH7 HK4+ HK2- 50 HK1 α-Gluc 45 α-Flag to DENV replication. fully described. Our team has already demonstrated that hepatitis C 40 80 80 HK2 α-Gluc *** 35 NS5A protein interacts and activates hexokinases (HKs) to favor viral 70 70 α-Flag cells 30 Poster presented at: presented Poster Fluorescente light Fluorescente 60 cells 60 α-Gluc replication [2]. It was described that dengue infection (DENV) 25 HK3 We observed that HuH7 HK4+HK2- cells have a rewiring of their 50 50 α-Flag 20 increases glycolysis [3] and thus we wondered if control of 40 40 glycolytic pathway resulting in intracellular lipids accumulation (see 15 HK4 α-Gluc 30 hexokinase activity was shared by DENV, another Flavivirus.