SUPPLEMENTARY DATA 1 Root Proteomic Analysis of Two Grapevine Rootstock Genotypes Showing Different Susceptibility to Salt Stres
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

METACYC ID Description A0AR23 GO:0004842 (Ubiquitin-Protein Ligase
Electronic Supplementary Material (ESI) for Integrative Biology This journal is © The Royal Society of Chemistry 2012 Heat Stress Responsive Zostera marina Genes, Southern Population (α=0. -

ATP-Citrate Lyase Has an Essential Role in Cytosolic Acetyl-Coa Production in Arabidopsis Beth Leann Fatland Iowa State University
Iowa State University Capstones, Theses and Retrospective Theses and Dissertations Dissertations 2002 ATP-citrate lyase has an essential role in cytosolic acetyl-CoA production in Arabidopsis Beth LeAnn Fatland Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/rtd Part of the Molecular Biology Commons, and the Plant Sciences Commons Recommended Citation Fatland, Beth LeAnn, "ATP-citrate lyase has an essential role in cytosolic acetyl-CoA production in Arabidopsis " (2002). Retrospective Theses and Dissertations. 1218. https://lib.dr.iastate.edu/rtd/1218 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Retrospective Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. ATP-citrate lyase has an essential role in cytosolic acetyl-CoA production in Arabidopsis by Beth LeAnn Fatland A dissertation submitted to the graduate faculty in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Major: Plant Physiology Program of Study Committee: Eve Syrkin Wurtele (Major Professor) James Colbert Harry Homer Basil Nikolau Martin Spalding Iowa State University Ames, Iowa 2002 UMI Number: 3158393 INFORMATION TO USERS The quality of this reproduction is dependent upon the quality of the copy submitted. Broken or indistinct print, colored or poor quality illustrations and photographs, print bleed-through, substandard margins, and improper alignment can adversely affect reproduction. In the unlikely event that the author did not send a complete manuscript and there are missing pages, these will be noted. -

Novel Insights Into Mannitol Metabolism in the Fungal Plant
Novel insights into mannitol metabolism in the fungal plant pathogen Botrytis cinerea Thierry Dulermo, Christine Rascle, Geneviève Billon-Grand, Elisabeth Gout, Richard Bligny, Pascale Cotton To cite this version: Thierry Dulermo, Christine Rascle, Geneviève Billon-Grand, Elisabeth Gout, Richard Bligny, et al.. Novel insights into mannitol metabolism in the fungal plant pathogen Botrytis cinerea. Biochemical Journal, Portland Press, 2010, 427 (2), pp.323-332. 10.1042/BJ20091813. hal-00479283 HAL Id: hal-00479283 https://hal.archives-ouvertes.fr/hal-00479283 Submitted on 30 Apr 2010 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Biochemical Journal Immediate Publication. Published on 05 Feb 2010 as manuscript BJ20091813 1 NOVEL INSIGHTS INTO MANNITOL METABOLISM IN THE FUNGAL PLANT 2 PATHOGEN BOTRYTIS CINEREA 3 4 Authors : Thierry Dulermo*†, Christine Rascle*, Geneviève Billon-Grand*, Elisabeth Gout‡, 5 Richard Bligny‡ and Pascale Cotton§ 6 7 Address 8 *Génomique Fonctionnelle des Champignons Pathogènes des Plantes, UMR 5240 9 Microbiologie, Adaptation -

Electronic Supplementary Material (ESI) for Molecular Biosystems
Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is © The Royal Society of Chemistry 2015 Table S5 Mass data of proteins identified with label free shotgun proteomics a #Alt. Proteins; b Scores; c #Peptides; d SC [%]; e RMS90 [ppm]; f Rank; g Median(Controls:GLP1); h #(Controls:GLP1); i CV [%](Controls:GLP1); l Median(Controls:Palmitate); m #(Controls:Palmitate) ; n CV [%](Controls:Palmitate); o Median(Controls:GLP1 + Palmitate); p #(Controls:GLP1 + Palmitate); q CV [%](Controls:GLP1 + Palmitate) MW OK Accession Protein pI a b c d e f g h i l m n o p q [kDa] 78 kDa glucose-regulated protein OS=Rattus 1672.6 true GRP78_RAT 72.3 4.9 1 22 39 1.48 1 1.16 6 20.53 1.18 9 9.56 1.24 10 13.52 norvegicus GN=Hspa5 PE=1 SV=1 (M:1672.6) Endoplasmin OS=Rattus norvegicus 1253.0 true ENPL_RAT 92.7 4.6 1 23 29 3.34 2 0.85 8 33.34 1.19 6 39.58 1.18 10 28.11 GN=Hsp90b1 PE=1 SV=2 (M:1253.0) Stress-70 protein, mitochondrial OS=Rattus 1138.8 true GRP75_RAT 73.8 5.9 1 17 28.7 1.18 3 1.21 5 7.62 0.78 1 1.1 8 8.04 norvegicus GN=Hspa9 PE=1 SV=3 (M:1138.8) Protein disulfide-isomerase A3 OS=Rattus 1035.4 true PDIA3_RAT 56.6 5.8 1 16 35.2 2.84 4 0.94 3 17.22 1.15 4 26.62 1.25 4 7.86 norvegicus GN=Pdia3 PE=1 SV=2 (M:1035.4) Aconitate hydratase, mitochondrial 983.8 true ACON_RAT OS=Rattus norvegicus GN=Aco2 PE=1 85.4 8.7 1 19 31.4 2.72 5 0.89 2 23.6 1.21 2 9.25 0.89 3 32.87 (M:983.8) SV=2 60 kDa heat shock protein, mitochondrial 906.9 true CH60_RAT OS=Rattus norvegicus GN=Hspd1 PE=1 60.9 5.8 1 13 32.5 3.26 6 1.05 5 13.15 1.01 2 32.54 1.01 -

Amino Mannitol Dehydrogenases on the Azasugar Biosynthetic Pathway
Send Orders for Reprints to [email protected] 10 Protein & Peptide Letters, 2014, 21, 10-14 Medium-Chain Dehydrogenases with New Specificity: Amino Mannitol Dehydrogenases on the Azasugar Biosynthetic Pathway Yanbin Wu, Jeffrey Arciola, and Nicole Horenstein* Department of Chemistry, University of Florida, Gainesville Florida, 32611-7200, USA Abstract: Azasugar biosynthesis involves a key dehydrogenase that oxidizes 2-amino-2-deoxy-D-mannitol to the 6-oxo compound. The genes encoding homologous NAD-dependent dehydrogenases from Bacillus amyloliquefaciens FZB42, B. atrophaeus 1942, and Paenibacillus polymyxa SC2 were codon-optimized and expressed in BL21(DE3) Escherichia coli. Relative to the two Bacillus enzymes, the enzyme from P. polymyxa proved to have superior catalytic properties with a Vmax of 0.095 ± 0.002 mol/min/mg, 59-fold higher than the B. amyloliquefaciens enzyme. The preferred substrate is 2- amino-2-deoxy-D-mannitol, though mannitol is accepted as a poor substrate at 3% of the relative rate. Simple amino alco- hols were also accepted as substrates at lower rates. Sequence alignment suggested D283 was involved in the enzyme’s specificity for aminopolyols. Point mutant D283N lost its amino specificity, accepting mannitol at 45% the rate observed for 2-amino-2-deoxy-D-mannitol. These results provide the first characterization of this class of zinc-dependent medium chain dehydrogenases that utilize aminopolyol substrates. Keywords: Aminopolyol, azasugar, biosynthesis, dehydrogenase, mannojirimycin, nojirimycin. INTRODUCTION are sufficient to convert fructose-6-phosphate into manno- jirimycin [9]. We proposed that the gutB1 gene product was Azasugars such as the nojirimycins [1] are natural prod- responsible for the turnover of 2-amino-2-deoxy-D-mannitol ucts that are analogs of monosaccharides that feature a nitro- (2AM) into mannojirimycin as shown in Fig. -

Molecular Mechanism for the Interaction Between Gibberellin and Brassinosteroid Signaling Pathways in Arabidopsis
Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis Javier Gallego-Bartoloméa, Eugenio G. Mingueta, Federico Grau-Enguixa, Mohamad Abbasa, Antonella Locascioa, Stephen G. Thomasb, David Alabadía,1, and Miguel A. Blázqueza aInstituto de Biología Molecular y Celular de Plantas, Consejo Superior de Investigaciones Científicas-Universidad Politécnica de Valencia, 46022 Valencia, Spain; and bRothamsted Research, Harpenden, Hertfordshire AL5 2JQ, United Kingdom Edited by Mark Estelle, University of California at San Diego, La Jolla, CA, and approved July 10, 2012 (received for review December 5, 2011) Plant development is modulated by the convergence of multiple response to auxin (10) and thus provides one of the possible environmental and endogenous signals, and the mechanisms that molecular mechanisms explaining the synergistic effect that both allow the integration of different signaling pathways is currently hormones exert on the expression of many genes (11). being unveiled. A paradigmatic case is the concurrence of brassinos- GAs and BRs regulate common physiological responses, e.g., teroid (BR) and gibberellin (GA) signaling in the control of cell expan- as illustrated by the dwarf phenotype of the GA- and BR-de- sion during photomorphogenesis, which is supported by phys- ficient mutants (6, 12). Moreover, both hormones act synergisti- iological observations in several plants but for which no molecular cally to promote hypocotyl elongation of light-grown Arabidopsis mechanism has been proposed. In this work, we show that the in- seedlings (13), a behavior that, as with BRs and auxin, might be tegration of these two signaling pathways occurs through the phys- interpreted as an indication of interaction between the two ical interaction between the DELLA protein GAI, which is a major pathways. -

Structures, Functions, and Mechanisms of Filament Forming Enzymes: a Renaissance of Enzyme Filamentation
Structures, Functions, and Mechanisms of Filament Forming Enzymes: A Renaissance of Enzyme Filamentation A Review By Chad K. Park & Nancy C. Horton Department of Molecular and Cellular Biology University of Arizona Tucson, AZ 85721 N. C. Horton ([email protected], ORCID: 0000-0003-2710-8284) C. K. Park ([email protected], ORCID: 0000-0003-1089-9091) Keywords: Enzyme, Regulation, DNA binding, Nuclease, Run-On Oligomerization, self-association 1 Abstract Filament formation by non-cytoskeletal enzymes has been known for decades, yet only relatively recently has its wide-spread role in enzyme regulation and biology come to be appreciated. This comprehensive review summarizes what is known for each enzyme confirmed to form filamentous structures in vitro, and for the many that are known only to form large self-assemblies within cells. For some enzymes, studies describing both the in vitro filamentous structures and cellular self-assembly formation are also known and described. Special attention is paid to the detailed structures of each type of enzyme filament, as well as the roles the structures play in enzyme regulation and in biology. Where it is known or hypothesized, the advantages conferred by enzyme filamentation are reviewed. Finally, the similarities, differences, and comparison to the SgrAI system are also highlighted. 2 Contents INTRODUCTION…………………………………………………………..4 STRUCTURALLY CHARACTERIZED ENZYME FILAMENTS…….5 Acetyl CoA Carboxylase (ACC)……………………………………………………………………5 Phosphofructokinase (PFK)……………………………………………………………………….6 -

Murine Neonatal Ketogenesis Preserves Mitochondrial Energetics by Preventing Protein Hyperacetylation
ARTICLES https://doi.org/10.1038/s42255-021-00342-6 Murine neonatal ketogenesis preserves mitochondrial energetics by preventing protein hyperacetylation Yuichiro Arima 1,2,13 ✉ , Yoshiko Nakagawa3,13, Toru Takeo 3,13, Toshifumi Ishida 1, Toshihiro Yamada1, Shinjiro Hino4, Mitsuyoshi Nakao4, Sanshiro Hanada 2, Terumasa Umemoto 2, Toshio Suda2, Tetsushi Sakuma 5, Takashi Yamamoto5, Takehisa Watanabe6, Katsuya Nagaoka6, Yasuhito Tanaka6, Yumiko K. Kawamura7,8, Kazuo Tonami7, Hiroki Kurihara7, Yoshifumi Sato9, Kazuya Yamagata9,10, Taishi Nakamura 1,11, Satoshi Araki1, Eiichiro Yamamoto1, Yasuhiro Izumiya1,12, Kenji Sakamoto1, Koichi Kaikita1, Kenichi Matsushita 1, Koichi Nishiyama2, Naomi Nakagata3 and Kenichi Tsujita1,10 Ketone bodies are generated in the liver and allow for the maintenance of systemic caloric and energy homeostasis during fasting and caloric restriction. It has previously been demonstrated that neonatal ketogenesis is activated independently of starvation. However, the role of ketogenesis during the perinatal period remains unclear. Here, we show that neonatal ketogen- esis plays a protective role in mitochondrial function. We generated a mouse model of insufficient ketogenesis by disrupting the rate-limiting hydroxymethylglutaryl-CoA synthase 2 enzyme gene (Hmgcs2). Hmgcs2 knockout (KO) neonates develop microvesicular steatosis within a few days of birth. Electron microscopic analysis and metabolite profiling indicate a restricted energy production capacity and accumulation of acetyl-CoA in Hmgcs2 KO mice. Furthermore, -
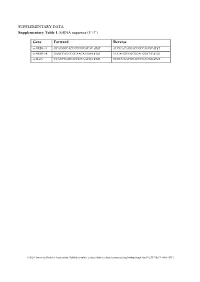
Supplementary Tables and Figures
SUPPLEMENTARY DATA Supplementary Table 1. SiRNA sequence (5’-3’) Gene Forward Reverse si-HRD1-1# GCAUGGCAGUCCUGUACAU dTdT AUGUACAGGACUGCCAUGC dTdT si-HRD1-2# GAGCCAUCCGCAACAUGAA dTdT UUCAUGUUGCGGAUGGCUC dTdT si-MafA CCAUCGAGUACGUCAACGA dTdT UCGUUGACGUACUCGAUGG dTdT ©2020 American Diabetes Association. Published online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db19-1060/-/DC1 SUPPLEMENTARY DATA Supplementary Table 2. Primer sequences for qRT-PCR (5’-3’) Gene Forward Reverse human HRD1 GCTCACGCCTACTACCTCAAA GCCAGACAAGTCTCTGTGACG mouse mafA AAGCGGCGCACGCTCAAGAA GGTCCCGCTCCTTGGCCAGA mouse insulin1 CACTTCCTACCCCTGCTGG ACCACAAAGATGCTGTTTGACA mouse β-actin AGGCCAACCGTGAAAAGATG AGAGCATAGCCCTCGTAGATGG human β-actin CATGTACGTTGCTATCCAGGC CTCCTTAATGTCACGCACGAT ©2020 American Diabetes Association. Published online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db19-1060/-/DC1 SUPPLEMENTARY DATA Supplementary Table 3. Primer sequences for ChIP (5’-3’) Gene promoter Forward Reverse mouse Insulin1, 2 GGAACTGTGAAACAGTCCAAGG CCCCCTGGACTTTGCTGTTTG ©2020 American Diabetes Association. Published online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db19-1060/-/DC1 SUPPLEMENTARY DATA Supplementary Table 4. Primer sequences for PCR (5’-3’) Gene Forward Reverse HRD1-pDsred CCCAAGCTTATGTTCCGCACCGCAGT GGGGTACCCAGTGGGCAACAGGGG HRD1-pCMV- Flag GGGGTACCATGTTCCGCACCGCAGT CCCAAGCTTGTGGGCAACAGGGGACT C HRD1-pCMV-HA GGCCATGGGCCATATGGGATCCTTCC AGGGATGCCACCCGGGGATCCTCAGT GCACCGCAGTGATG GGGCAACAGGGGAC HRD1-N-HA GGCCATGGGCCATATGGGATCCTTCC -

Integration of Auxin and Brassinosteroid Pathways by Auxin Response Factor 2
Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2 Gre´ gory Vert†, Cristina L. Walcher‡, Joanne Chory§¶, and Jennifer L. Nemhauser‡¶ †Biochimie and Physiologie Mole´culaire des Plantes, Centre National de la Recherche Scientifique, Unite´Mixte de Recherche 5004, Institut de Biologie Inte´grative des Plantes, 2 Place Viala, 34060 Montpellier Cedex 1, France; ‡Department of Biology, University of Washington, Box 351800, Seattle, WA 98195-1800; and §Howard Hughes Medical Institute and Plant Biology Laboratory, The Salk Institute, 10010 North Torrey Pines Road, La Jolla, CA 92037 Contributed by Joanne Chory, April 25, 2008 (sent for review March 5, 2008) Plant form is shaped by a complex network of intrinsic and extrinsic bind and activate transcription on the promoters of genes like signals. Light-directed growth of seedlings (photomorphogenesis) SAUR-15, common to both auxin and BR pathways (13). BRs depends on the coordination of several hormone signals, including regulate the activity of BES1 and BZR1 by modulating their brassinosteroids (BRs) and auxin. Although the close relationship phosphorylation status, dependent on the coordinate action of between BRs and auxin has been widely reported, the molecular the BIN2 GSK3 kinase and the BSU1 phosphatase, and respec- mechanism for combinatorial control of shared target genes has tive family members (7, 15–18). Phosphorylation by BIN2 in- remained elusive. Here we demonstrate that BRs synergistically hibits homodimerization by BES1, thereby blocking DNA- increase seedling sensitivity to auxin and show that combined binding and transcriptional activation (17), and modulates the treatment with both hormones can increase the magnitude and dynamics of BZR1 in the cell by interaction with a 14-3-3 protein duration of gene expression. -

PIN-LIKES Coordinate Brassinosteroid Signalling with Nuclear Auxin Input in Arabidopsis Thaliana
bioRxiv preprint doi: https://doi.org/10.1101/646489; this version posted July 19, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. PIN-LIKES coordinate brassinosteroid signalling with nuclear auxin input in Arabidopsis thaliana Authors: Lin Sun1, Elena Feraru1, Mugurel I. Feraru1, Krzysztof Wabnik2, Jürgen Kleine-Vehn1,* Affiliations: 1Department of Applied Genetics and Cell Biology, University of Natural Resources and Life Sciences (BOKU), Muthgasse 18, 1190 Vienna, Austria 2Centro de Biotecnología y Genómica de Plantas (Universidad Politécnica de Madrid - Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria), Autopista M-40, Km 38 - 28223 Pozuelo de Alarcón, Spain *Correspondence should be addressed to J.K.-V. ([email protected]) 1 bioRxiv preprint doi: https://doi.org/10.1101/646489; this version posted July 19, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Auxin and brassinosteroids (BR) are crucial growth regulators and display overlapping functions during plant development. Here, we reveal an alternative phytohormone crosstalk mechanism, revealing that brassinosteroid signaling controls nuclear abundance of auxin. We performed a forward genetic screen for imperial pils (imp) mutants that enhance the overexpression phenotypes of PIN-LIKES (PILS) putative intracellular auxin transport facilitator. Here we report that the imp1 mutant is defective in the brassinosteroid-receptor BRI1. Our data reveals that BR signaling transcriptionally and posttranslationally represses accumulation of PILS proteins at the endoplasmic reticulum, thereby increasing nuclear abundance and signaling of auxin. -

Genetic Characterisaton of Rhodococcus Rhodochrous ATCC
The copyright of this thesis vests in the author. No quotation from it or information derived from it is to be published without full acknowledgement of the source. The thesis is to be used for private study or non- commercial research purposes only. Published by the University of Cape Town (UCT) in terms of the non-exclusive license granted to UCT by the author. University of Cape Town Genetic characterization of Rhodococcus rhodochrous ATCC BAA-870 with emphasis on nitrile hydrolysing enzymes n ow Joni Frederick A thesis submitted in fulfilment of the requirements for the degree of Doctor of Philosophy in the Departmentty of of MolecularCape and T Cell Biology, Universitysi of Cape Town er UnivSupervisor: Professor B. T. Sewell Co-supervisor: Professor D. Brady February 2013 Keywords Nitrile hydrolysis Biocatalysis Rhodococcus rhodochrous ATCC BAA-870 Genome sequencing Nitrilase Nitrile hydratase n ow ty of Cape T si er Univ ii Keywords Abstract Rhodococcus rhodochrous ATCC BAA-870 (BAA-870) had previously been isolated on selective media for enrichment of nitrile hydrolysing bacteria. The organism was found to have a wide substrate range, with activity against aliphatics, aromatics, and aryl aliphatics, and enantioselectivity towards beta substituted nitriles and beta amino nitriles, compounds that have potential applications in the pharmaceutical industry. This makes R. rhodochrous ATCC BAA-870 potentially a versatile biocatalyst for the synthesis of a broad range of compounds with amide and carboxylic acid groups that can be derived from structurally related nitrile precursors. The selectivity of biocatalysts allows for high product yields and better atom economyn than non- selective chemical methods of performing this reaction, suchow as acid or base hydrolysis.