Dectin-1 Limits Central Nervous System Autoimmunity Through a Non-Canonical Pathway
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Microglial Function and Regulation During Development, Homeostasis and Alzheimer’S Disease
cells Review Microglial Function and Regulation during Development, Homeostasis and Alzheimer’s Disease Brad T. Casali and Erin G. Reed-Geaghan * Department of Pharmaceutical Sciences, Northeast Ohio Medical University, Rootstown, OH 44272, USA; [email protected] * Correspondence: [email protected] Abstract: Microglia are the resident immune cells of the brain, deriving from yolk sac progenitors that populate the brain parenchyma during development. During development and homeostasis, microglia play critical roles in synaptogenesis and synaptic plasticity, in addition to their primary role as immune sentinels. In aging and neurodegenerative diseases generally, and Alzheimer’s disease (AD) specifically, microglial function is altered in ways that significantly diverge from their homeostatic state, inducing a more detrimental inflammatory environment. In this review, we discuss the receptors, signaling, regulation and gene expression patterns of microglia that mediate their phenotype and function contributing to the inflammatory milieu of the AD brain, as well as strategies that target microglia to ameliorate the onset, progression and symptoms of AD. Keywords: microglia; inflammation; Alzheimer’s disease; neurodegenerative diseases; TREM2; neu- roinflammation Citation: Casali, B.T.; Reed-Geaghan, 1. Introduction E.G. Microglial Function and Microglia are the resident phagocytes of the central nervous system (CNS). In addition Regulation during Development, to their immunological role in maintaining CNS homeostasis, microglia play vital -

Card9 As a Critical Regulator of Tumor Development
Cancer Letters 451 (2019) 150–155 Contents lists available at ScienceDirect Cancer Letters journal homepage: www.elsevier.com/locate/canlet Mini-review Card9 as a critical regulator of tumor development T ∗ Xiaoming Zhongb,1, Bin Chenc,1, Liang Yangd, Zhiwen Yanga, a Department of Pharmacy, Songjiang Hospital Affiliated Shanghai First People's Hospital, Shanghai Jiao Tong University, Shanghai, China b Jiangxi Province Tumor Hospital, Nanchang, China c Surgery Department, First Affiliated Hospital of Gannan Medical University, Gannan Medical University, Ganzhou, China d Nanjing Medical University, The Affiliated Changzhou No.2 People's Hospital, Nanjing Medical University, Nanjing, China ARTICLE INFO ABSTRACT Keywords: Caspase recruitment domain-containing protein 9 (Card9) is a myeloid cell-specific signaling protein that plays a Card9 critical role in NF-κB and MAPK activation. This leads to initiation of the inflammatory cytokine cascade, and Macrophages elicits the host immune response against microbial invasion, especially in fungal infection. Current research Tumor growth indicates that Card9 plays an important role in tumor progression. Here, we review the data from preclinical and Target therapy clinical studies of Card9 and suggest the potential for Card9-targeted interventions in the prevention or treat- ment of certain tumors. 1. Introduction humans. Among the various etiological factors, hepatitis C virus (HCV) infection is a major cause of HCC [11]. It is of great value for under- Caspase recruitment domain-containing protein 9 (Card9) is a cen- standing the HCC progression in HCV-infected patients. tral integrator of innate and adaptive immunity that is mainly expressed Zekri and colleagues studied 130 patients with HCV-associated liver in myeloid cells, especially in macrophages and dendritic cells. -
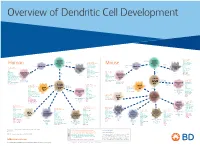
View Dendritic Cell Development Poster
Overview of Dendritic Cell Development Lineage–, CD45+, Common CD117 (c-kit) Common MHCII+, CD11c+ – + CD207 (Langerin) Myeloid CD117 (c-kit) Lineage , CD45 , Myeloid Progenitor MHCII (HLA-DR)+, CD11c+ Progenitor CD324 (E-Cadherin) Human Mouse CD326 (EpCAM) CD207 (Langerin) TGFb1 Cells CD11b, CD115 Cells CD14 Monocyte CD324 (E-Cadherin) Monocyte M-CSF CD11b – + Ly6C Langerhans CD24 Lineage , CD45 , M-CSF CD326 (EpCAM) MHCII (HLA-DR)+, CD11c+ Langerhans CD11blo Zbtb46– Cells CD172a (Sirp-α) CD16 CD1ahi, CD1c CD205 (DEC-205) Cells CSF F4/80 CD64 CD172a (Sirp-α) Lineage–, CD45+, FLT3L TLR3, TLR11 CD1a, CD1c Inflammatory CD369 (Dectin-1/CLEC7A) MHCII+, CD11c+ +/– CSF IL-15 CD8–, CD14– CD11b, CD14 CD371 (CLEC12A) CD64 Monocyte- FLT3L Inflammatory CD370 (Clec9a)– CD172a (Sirp-α) IL-15 CLEC6A CD11b derived lo Monocyte- CD206, CD209 (DC-SIGN) TLR1, TLR2, TLR3 , TLR6 CD209a (DC-SIGN) CD367 (DCIR/CLEC4A) DCs CD14– CD272 (BTLA)lo derived CD369 (Dectin-1/CLEC7A) DCs Common Ly-6C – + CD371 (CLEC12A) CD117 (c-kit) Lineage , CD45 , IL-1β, IL-6, IL-10, TLR1-6, TLR7-8, TLR10 Dendritic + lo CLEC6A – – CD135/FLT3 MHCII , CD11c IL-12, IL-23, TNF CD8a , CD14 IL-1β, IL-6 IL-10, Precursor TLR3lo, TLR4, TLR7, TLR8 CD45R (B220) IL-12, IL-23, TNF Plasmacytoid CD207 (Langerin)– Cells CD317 (BST-2) Common Lineage–, CD45+, FLT3L DCs Lineage–, CD45+, + Ly6C + lo/– CD207 IFN Type I + + Dendritic CD135/FLT3 MHCII (HLA-DR) , CD11c Lineage–, CD45+, IRF7, IRF8, BATF3hi Siglec-H MHCII (HLA-DR) , CD11c hi – + CD123 + + Dermal SpiB, Zbtb46 CD1a, CD64 CD1a Precursor CD117 (c-kit) -

Human Lectins, Their Carbohydrate Affinities and Where to Find Them
biomolecules Review Human Lectins, Their Carbohydrate Affinities and Where to Review HumanFind Them Lectins, Their Carbohydrate Affinities and Where to FindCláudia ThemD. Raposo 1,*, André B. Canelas 2 and M. Teresa Barros 1 1, 2 1 Cláudia D. Raposo * , Andr1 é LAQVB. Canelas‐Requimte,and Department M. Teresa of Chemistry, Barros NOVA School of Science and Technology, Universidade NOVA de Lisboa, 2829‐516 Caparica, Portugal; [email protected] 12 GlanbiaLAQV-Requimte,‐AgriChemWhey, Department Lisheen of Chemistry, Mine, Killoran, NOVA Moyne, School E41 of ScienceR622 Co. and Tipperary, Technology, Ireland; canelas‐ [email protected] NOVA de Lisboa, 2829-516 Caparica, Portugal; [email protected] 2* Correspondence:Glanbia-AgriChemWhey, [email protected]; Lisheen Mine, Tel.: Killoran, +351‐212948550 Moyne, E41 R622 Tipperary, Ireland; [email protected] * Correspondence: [email protected]; Tel.: +351-212948550 Abstract: Lectins are a class of proteins responsible for several biological roles such as cell‐cell in‐ Abstract:teractions,Lectins signaling are pathways, a class of and proteins several responsible innate immune for several responses biological against roles pathogens. such as Since cell-cell lec‐ interactions,tins are able signalingto bind to pathways, carbohydrates, and several they can innate be a immuneviable target responses for targeted against drug pathogens. delivery Since sys‐ lectinstems. In are fact, able several to bind lectins to carbohydrates, were approved they by canFood be and a viable Drug targetAdministration for targeted for drugthat purpose. delivery systems.Information In fact, about several specific lectins carbohydrate were approved recognition by Food by andlectin Drug receptors Administration was gathered for that herein, purpose. plus Informationthe specific organs about specific where those carbohydrate lectins can recognition be found by within lectin the receptors human was body. -

A New Vision of Iga Nephropathy: the Missing Link
International Journal of Molecular Sciences Review A New Vision of IgA Nephropathy: The Missing Link Fabio Sallustio 1,2,* , Claudia Curci 2,3,* , Vincenzo Di Leo 3 , Anna Gallone 2, Francesco Pesce 3 and Loreto Gesualdo 3 1 Interdisciplinary Department of Medicine (DIM), University of Bari “Aldo Moro”, 70124 Bari, Italy 2 Department of Basic Medical Sciences, Neuroscience and Sense Organs, University of Bari “Aldo Moro”, 70124 Bari, Italy; [email protected] 3 Nephrology, Dialysis and Transplantation Unit, DETO, University “Aldo Moro”, 70124 Bari, Italy; [email protected] (V.D.L.); [email protected] (F.P.); [email protected] (L.G.) * Correspondence: [email protected] (F.S.); [email protected] (C.C.) Received: 7 December 2019; Accepted: 24 December 2019; Published: 26 December 2019 Abstract: IgA Nephropathy (IgAN) is a primary glomerulonephritis problem worldwide that develops mainly in the 2nd and 3rd decade of life and reaches end-stage kidney disease after 20 years from the biopsy-proven diagnosis, implying a great socio-economic burden. IgAN may occur in a sporadic or familial form. Studies on familial IgAN have shown that 66% of asymptomatic relatives carry immunological defects such as high IgA serum levels, abnormal spontaneous in vitro production of IgA from peripheral blood mononuclear cells (PBMCs), high serum levels of aberrantly glycosylated IgA1, and an altered PBMC cytokine production profile. Recent findings led us to focus our attention on a new perspective to study the pathogenesis of this disease, and new studies showed the involvement of factors driven by environment, lifestyle or diet that could affect the disease. -

Defining the Relevant Combinatorial Space of the PKC/CARD-CC Signal Transduction Nodes
bioRxiv preprint doi: https://doi.org/10.1101/228767; this version posted May 2, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Defining the relevant combinatorial space of the PKC/CARD-CC signal transduction nodes Jens Staal1,2,*, Yasmine Driege1,2, Mira Haegman1,2, Styliani Iliaki1,2, Domien Vanneste1,2, Inna Affonina1,2, Harald Braun1,2, Rudi Beyaert1,2 1 Department of Biomedical Molecular Biology, Ghent University, Ghent, Belgium, 2 VIB-UGent Center for Inflammation Research, Unit of Molecular Signal Transduction in Inflammation, VIB, Ghent, Belgium. * corresponding author: [email protected] Running title: PKC/CARD-CC signaling Abbreviations: Bcl10 = B Cell CLL/Lymphoma 10 CARD = Caspase activation and recruitment domain CC = Coiled-coil domain MALT1 = Mucosa-associated lymphoid tissue lymphoma translocation protein 1 PKC = protein kinase C Keywords: Inflammation, cancer, NF-kappaB, paracaspase Conflict of interests: The authors declare no conflict of interest. bioRxiv preprint doi: https://doi.org/10.1101/228767; this version posted May 2, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Abstract Biological signal transduction typically display a so-called bow-tie or hour glass topology: Multiple receptors lead to multiple cellular responses but the signals all pass through a narrow waist of central signaling nodes. -

Immunology Focus Summer | 2006 Immunology FOCUS
R&D Systems Immunology Focus summer | 2006 Immunology FOCUS Inside page 2 Signal Transduction: Kinase & Phosphatase Reagents page 3 Lectin Family page 4 Regulatory T Cells page 5 Natural Killer Cells page 6 Innate Immunity & Dendritic Cells page 7 Co-Stimulation/-Inhibition The B7 Family & Associated Molecules page 8 Proteome Profiler™ Phospho-Immunoreceptor Array ITAM/ITIM-Associated Receptors www.RnDSystems.com Please visit our website @ www.RnDSystems.com for product information and past issues of the Focus Newsletter: Cancer, Neuroscience, Cell Biology, and more. Quality | Selec tion | Pe rformance | Result s Cancer Development Endocrinology Immunology Neuroscience Proteases Stem Cells Signal Transduction: Kinase & Phosphatase Reagents Co-inhibitory PD-L2/PD-1 signaling & SHP-2 phosphatase KINASE & PHOSPHATASE RESEARCH REAGENTS Regulation of MAP kinase (MAPK) signaling Kinases Phosphatases pathways is critical for T cell development, MOLECULE ANTIBODIES ELISAs/ASSAYS MOLECULE ANTIBODIES ELISAs/ASSAYS activation, differentiation, and death. MAPKs Akt Family H M R H M R Alkaline Phosphatase* H M R are activated by the dual phosphorylation of threonine and tyrosine residues resulting in AMPK H M R Calcineurin A, B H M R subsequent transcription factor activation. ATM H M R H CD45 H M H M The MAPK signaling pathway in T cells can be CaM Kinase II Ms CDC25A, B* H M R triggered by cytokines, growth factors, and CDC2 H M R DARPP-32 M R ligands for transmembrane receptors. Chk1, 2 H M R H M R DEP-1/CD148* H M R H Ligation of the T cell receptor (TCR)/CD3 ERK1, 2 H M R H M R LAR H M R complex results in rapid activation of PI 3- kinase, which leads to Akt and MAPK ERK3 H Lyp H activation. -

Type I Interferon Response Drives Neuroinflammation and Synapse Loss in Alzheimer Disease
RESEARCH ARTICLE The Journal of Clinical Investigation Type I interferon response drives neuroinflammation and synapse loss in Alzheimer disease Ethan R. Roy,1,2 Baiping Wang,1 Ying-wooi Wan,3 Gabriel Chiu,1 Allysa Cole,1 Zhuoran Yin,4 Nicholas E. Propson,1,5 Yin Xu,1 Joanna L. Jankowsky,6 Zhandong Liu,7 Virginia M.-Y. Lee,8 John Q. Trojanowski,8 Stephen D. Ginsberg,9,10 Oleg Butovsky,4,11 Hui Zheng,1,3 and Wei Cao1,3 1Huffington Center on Aging, 2Translational Biology & Molecular Medicine Program, and 3Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas, USA. 4Ann Romney Center for Neurologic Diseases, Department of Neurology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA. 5Molecular and Cellular Biology Program, Department of Molecular and Cellular Biology, 6Department of Neuroscience, and 7Department of Pediatrics-Neurology, Baylor College of Medicine, Houston, Texas, USA. 8Department of Pathology and Laboratory Medicine, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania, USA. 9Center for Dementia Research, Nathan Kline Institute, Orangeburg, New York, USA. 10Departments of Psychiatry, Neuroscience & Physiology and the NYU Neuroscience Institute, New York University Langone Medical Center, New York, New York, USA. 11Evergrande Center for Immunologic Diseases, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA. Type I interferon (IFN) is a key cytokine that curbs viral infection and cell malignancy. Previously, we demonstrated a potent IFN immunogenicity of nucleic acid–containing (NA-containing) amyloid fibrils in the periphery. Here, we investigated whether IFN is associated with β-amyloidosis inside the brain and contributes to neuropathology. -

Influence of Galectin-3 on the Innate Immune Response During
Journal of Fungi Article Influence of Galectin-3 on the Innate Immune Response during Experimental Cryptococcosis Caroline Patini Rezende 1, Patricia Kellen Martins Oliveira Brito 2 , Thiago Aparecido Da Silva 2 , Andre Moreira Pessoni 1 , Leandra Naira Zambelli Ramalho 3 and Fausto Almeida 1,* 1 Department of Biochemistry and Immunology, Ribeirao Preto Medical School, University of Sao Paulo, Ribeirao Preto 14049-900, SP, Brazil; [email protected] (C.P.R.); [email protected] (A.M.P.) 2 Department of Cellular and Molecular Biology, Ribeirao Preto Medical School, University of Sao Paulo, Ribeirao Preto 14049-900, SP, Brazil; [email protected] (P.K.M.O.B.); [email protected] (T.A.D.S.) 3 Department of Pathology, Ribeirao Preto Medical School, University of Sao Paulo, Ribeirao Preto 14049-900, SP, Brazil; [email protected] * Correspondence: [email protected] Abstract: Cryptococcus neoformans, the causative agent of cryptococcosis, is the primary fungal pathogen that affects the immunocompromised individuals. Galectin-3 (Gal-3) is an animal lectin involved in both innate and adaptive immune responses. The present study aimed to evaluate the influence of Gal-3 on the C. neoformans infection. We performed histopathological and gene profile analysis of the innate antifungal immunity markers in the lungs, spleen, and brain of the wild-type (WT) and Gal-3 knockout (KO) mice during cryptococcosis. These findings suggest that Gal-3 absence does not cause significant histopathological alterations in the analyzed tissues. The expression profile of the genes related to innate antifungal immunity showed that the presence of cryptococcosis in Citation: Rezende, C.P.; Brito, the WT and Gal-3 KO animals, compared to their respective controls, promoted the upregulation P.K.M.O.; Da Silva, T.A.; Pessoni, of the pattern recognition receptor (PRR) responsive to mannose/chitin (mrc1) and a gene involved A.M.; Ramalho, L.N.Z.; Almeida, F. -

Small-Molecule Inhibitors Directly Target CARD9 and Mimic Its Protective Variant in Inflammatory Bowel Disease
Small-molecule inhibitors directly target CARD9 and mimic its protective variant in inflammatory bowel disease Elizaveta S. Leshchinera,b, Jason S. Rushc, Michael A. Durneyc, Zhifang Caod,e,f, Vlado Dancíkˇ b, Benjamin Chittickb, Huixian Wub, Adam Petronec, Joshua A. Bittkerc, Andrew Phillipsc, Jose R. Perezc, Alykhan F. Shamjib, Virendar K. Kaushikc, Mark J. Dalyg,h, Daniel B. Grahamd,e,f, Stuart L. Schreibera,b,1, and Ramnik J. Xavierd,e,f,1 aDepartment of Chemistry and Chemical Biology, Harvard University, Cambridge, MA 02138; bCenter for the Science of Therapeutics, Broad Institute, Cambridge, MA 02142; cCenter for the Development of Therapeutics, Broad Institute, Cambridge, MA 02142; dGastrointestinal Unit, Massachusetts General Hospital, Harvard Medical School, Boston, MA 02114; eCenter for the Study of Inflammatory Bowel Disease, Massachusetts General Hospital, Harvard Medical School, Boston, MA 02114; fInfectious Disease and Microbiome Program, Broad Institute, Cambridge, MA 02142; gMedical and Population Genetics Program, Broad Institute, Cambridge, MA 02142; and hAnalytic and Translational Genetics Unit, Massachusetts General Hospital, Boston, MA 02114 Contributed by Stuart L. Schreiber, September 7, 2017 (sent for review April 19, 2017; reviewed by Benjamin F. Cravatt and Kevan M. Shokat) Advances in human genetics have dramatically expanded our the gap between genetic knowledge in IBD and its therapeutic understanding of complex heritable diseases. Genome-wide associ- potential by focusing on protective variants that both reveal the ation studies have identified an allelic series of CARD9 variants as- mechanisms of disease pathogenesis and suggest safe and effec- sociated with increased risk of or protection from inflammatory tive therapeutic strategies. bowel disease (IBD). -

Immunity Against Fungi
Immunity against fungi Michail S. Lionakis, … , Iliyan D. Iliev, Tobias M. Hohl JCI Insight. 2017;2(11):e93156. https://doi.org/10.1172/jci.insight.93156. Review Pathogenic fungi cause a wide range of syndromes in immune-competent and immune-compromised individuals, with life-threatening disease primarily seen in humans with HIV/AIDS and in patients receiving immunosuppressive therapies for cancer, autoimmunity, and end-organ failure. The discovery that specific primary immune deficiencies manifest with fungal infections and the development of animal models of mucosal and invasive mycoses have facilitated insight into fungus-specific recognition, signaling, effector pathways, and adaptive immune responses. Progress in deciphering the molecular and cellular basis of immunity against fungi is guiding preclinical studies into vaccine and immune reconstitution strategies for vulnerable patient groups. Furthermore, recent work has begun to address the role of endogenous fungal communities in human health and disease. In this review, we summarize a contemporary understanding of protective immunity against fungi. Find the latest version: https://jci.me/93156/pdf REVIEW Immunity against fungi Michail S. Lionakis,1 Iliyan D. Iliev,2 and Tobias M. Hohl3 1Fungal Pathogenesis Unit, Laboratory of Clinical Infectious Diseases, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, Maryland, USA. 2Jill Roberts Institute for Research in IBD, Department of Medicine, Weill Cornell Medical College, New York, New York, USA. 3Infectious Disease Service, Department of Medicine, and Immunology Program, Memorial Sloan Kettering Cancer Center, New York, New York, USA. Pathogenic fungi cause a wide range of syndromes in immune-competent and immune- compromised individuals, with life-threatening disease primarily seen in humans with HIV/AIDS and in patients receiving immunosuppressive therapies for cancer, autoimmunity, and end-organ failure. -

Growth Faltering Regardless of Chronic Diarrhea Is Associated with Mucosal Immune Dysfunction and Microbial Dysbiosis in the Gut Lumen
www.nature.com/mi ARTICLE OPEN Growth faltering regardless of chronic diarrhea is associated with mucosal immune dysfunction and microbial dysbiosis in the gut lumen 1 2 3 3 4 5 4 Nicholas S. Rhoades , Sara M. Hendrickson✉ , Kamm Prongay , Andrew Haertel , Leanne Gill , Robert A. Edwards , Laura Garzel , Mark K. Slifka2 and Ilhem Messaoudi1 © The Author(s) 2021 Despite the impact of childhood diarrhea on morbidity and mortality, our understanding of its sequelae has been significantly hampered by the lack of studies that examine samples across the entire intestinal tract. Infant rhesus macaques are naturally susceptible to human enteric pathogens and recapitulate the hallmarks of diarrheal disease such as intestinal inflammation and growth faltering. Here, we examined intestinal biopsies, lamina propria leukocytes, luminal contents, and fecal samples from healthy infants and those experiencing growth faltering with distant acute or chronic active diarrhea. We show that growth faltering in the presence or absence of active diarrhea is associated with a heightened systemic and mucosal pro-inflammatory state centered in the colon. Moreover, polyclonal stimulation of colonic lamina propria leukocytes resulted in a dampened cytokine response, indicative of immune exhaustion. We also detected a functional and taxonomic shift in the luminal microbiome across multiple gut sites including the migration of Streptococcus and Prevotella species between the small and large intestine, suggesting a decompartmentalization of gut microbial communities. Our studies provide valuable insight into the outcomes of diarrheal diseases and growth faltering not attainable in humans and lays the groundwork to test interventions in a controlled and reproducible setting. Mucosal Immunology (2021) 14:1113–1126; https://doi.org/10.1038/s41385-021-00418-2 INTRODUCTION especially in the genus Campylobacter, that are missed by clinical Despite significant improvements in healthcare delivery and tests but highly abundant in developing countries24–26.