Influence of Galectin-3 on the Innate Immune Response During
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Idiopathic CD4 Lymphocytopenia: Spectrum of Opportunistic Infections, Malignancies, and Autoimmune Diseases
Published online: 2021-08-09 REVIEW ARTICLE Idiopathic CD4 Lymphocytopenia: Spectrum of opportunistic infections, malignancies, and autoimmune diseases Dina S. Ahmad, Mohammad Esmadi, William C. Steinmann Department of Internal Medicine, University of Missouri School of Medicine, Columbia, MO, USA Access this article online ABSTRACT Website: www.avicennajmed.com DOI: 10.4103/2231-0770.114121 Idiopathic CD4 lymphocytopenia (ICL) was first defined in 1992 by the US Centers for Disease Quick Response Code: Control and Prevention (CDC) as the repeated presence of a CD4+ T lymphocyte count of fewer than 300 cells per cubic millimeter or of less than 20% of total T cells with no evidence of human immunodeficiency virus (HIV) infection and no condition that might cause depressed CD4 counts. Most of our knowledge about ICL comes from scattered case reports. The aim of this study was to collect comprehensive data from the previously published cases to understand the characteristics of this rare condition. We searched the PubMed database and Science Direct for case reports since 1989 for Idiopathic CD4 lymphocytopenia cases. We found 258 cases diagnosed with ICL in 143 published papers. We collected data about age, sex, pathogens, site of infections, CD4 count, CD8 count, CD4:CD8 ratio, presence of HIV risk factors, malignancies, autoimmune diseases and whether the patients survived or died. The mean age at diagnosis of first opportunistic infection (or ICL if no opportunistic infection reported) was 40.7 ± 19.2 years (standard deviation), with a range of 1 to 85. One-sixty (62%) patients were males, 91 (35.2%) were females, and 7 (2.7%) patients were not identified whether males or females. -

153.Full.Pdf
Copyright Ó 2010 by the Genetics Society of America DOI: 10.1534/genetics.109.113027 A Tetrad Analysis of the Basidiomycete Fungus Cryptococcus neoformans Alexander Idnurm Division of Cell Biology and Biophysics, School of Biological Sciences, University of Missouri-Kansas City, Kansas City, Missouri 64110 Manuscript received December 10, 2009 Accepted for publication February 9, 2010 ABSTRACT Cryptococcus neoformans is a basidiomycete fungus that is found worldwide and causes disease in humans and animal species. The fungus grows asexually as a budding yeast. Under laboratory conditions it is capable of sexual reproduction between two mating types. After cell fusion a dikaryotic filament develops, at the tip of which a basidium gives rise to four chains of basidiospores. Because the chains each comprise 10–30 spores, rather than single spores, the analysis of individual meiotic events has not been attempted in C. neoformans in the style of tetrad analyses performed in other fungal species. Here, the basidiospores from .100 basidia were micromanipulated and the resultant .2500 progeny analyzed for three genetic markers to understand the sexual process in this fungus, leading to four observations: (i) Marker seg- regation provides genetic evidence for a single meiotic event within the basidium followed by multiple rounds of mitosis. (ii) Using each basidium as an unordered tetrad, the ADE2 and URA5 genes are linked to their centromeres, consistent with adjacent genomic regions rich in repetitive elements predicted to comprise Cryptococcus centromeres. (iii) Lack of germination of basidiospores is attributed to aneuploidy, rather than dormancy. (iv) Analysis of basidiospores derived from single chains demonstrates that each chain can contain different genotypes. -

Microglial Function and Regulation During Development, Homeostasis and Alzheimer’S Disease
cells Review Microglial Function and Regulation during Development, Homeostasis and Alzheimer’s Disease Brad T. Casali and Erin G. Reed-Geaghan * Department of Pharmaceutical Sciences, Northeast Ohio Medical University, Rootstown, OH 44272, USA; [email protected] * Correspondence: [email protected] Abstract: Microglia are the resident immune cells of the brain, deriving from yolk sac progenitors that populate the brain parenchyma during development. During development and homeostasis, microglia play critical roles in synaptogenesis and synaptic plasticity, in addition to their primary role as immune sentinels. In aging and neurodegenerative diseases generally, and Alzheimer’s disease (AD) specifically, microglial function is altered in ways that significantly diverge from their homeostatic state, inducing a more detrimental inflammatory environment. In this review, we discuss the receptors, signaling, regulation and gene expression patterns of microglia that mediate their phenotype and function contributing to the inflammatory milieu of the AD brain, as well as strategies that target microglia to ameliorate the onset, progression and symptoms of AD. Keywords: microglia; inflammation; Alzheimer’s disease; neurodegenerative diseases; TREM2; neu- roinflammation Citation: Casali, B.T.; Reed-Geaghan, 1. Introduction E.G. Microglial Function and Microglia are the resident phagocytes of the central nervous system (CNS). In addition Regulation during Development, to their immunological role in maintaining CNS homeostasis, microglia play vital -

Card9 As a Critical Regulator of Tumor Development
Cancer Letters 451 (2019) 150–155 Contents lists available at ScienceDirect Cancer Letters journal homepage: www.elsevier.com/locate/canlet Mini-review Card9 as a critical regulator of tumor development T ∗ Xiaoming Zhongb,1, Bin Chenc,1, Liang Yangd, Zhiwen Yanga, a Department of Pharmacy, Songjiang Hospital Affiliated Shanghai First People's Hospital, Shanghai Jiao Tong University, Shanghai, China b Jiangxi Province Tumor Hospital, Nanchang, China c Surgery Department, First Affiliated Hospital of Gannan Medical University, Gannan Medical University, Ganzhou, China d Nanjing Medical University, The Affiliated Changzhou No.2 People's Hospital, Nanjing Medical University, Nanjing, China ARTICLE INFO ABSTRACT Keywords: Caspase recruitment domain-containing protein 9 (Card9) is a myeloid cell-specific signaling protein that plays a Card9 critical role in NF-κB and MAPK activation. This leads to initiation of the inflammatory cytokine cascade, and Macrophages elicits the host immune response against microbial invasion, especially in fungal infection. Current research Tumor growth indicates that Card9 plays an important role in tumor progression. Here, we review the data from preclinical and Target therapy clinical studies of Card9 and suggest the potential for Card9-targeted interventions in the prevention or treat- ment of certain tumors. 1. Introduction humans. Among the various etiological factors, hepatitis C virus (HCV) infection is a major cause of HCC [11]. It is of great value for under- Caspase recruitment domain-containing protein 9 (Card9) is a cen- standing the HCC progression in HCV-infected patients. tral integrator of innate and adaptive immunity that is mainly expressed Zekri and colleagues studied 130 patients with HCV-associated liver in myeloid cells, especially in macrophages and dendritic cells. -

Product Sheet Info
Product Information Sheet for NR-50430 Cryptococcus gattii, Strain C10 Propagation: 1. Keep vial frozen until ready for use; thaw rapidly. Catalog No. NR-50430 2. Inoculate an agar plate with approximately 50 µL of thawed culture and/or transfer the entire thawed aliquot into a single tube of broth For research use only. Not for human use. 3. Incubate the plate and/or tube at 25°C for 2 to 4 days. Contributor: Citation: Brian Wong, M.D., Professor, and Igor Bruzual, Ph.D., Acknowledgment for publications should read “The following Infectious Disease Division, Department of Medicine, Oregon reagent was obtained through BEI Resources, NIAID, NIH: Health and Science University, Portland, Oregon, USA Cryptococcus gattii, Strain C10, NR-50430.” Manufacturer: Biosafety Level: 2 BEI Resources Appropriate safety procedures should always be used with this material. Laboratory safety is discussed in the following Product Description: publication: U.S. Department of Health and Human Services, Classification: Tremellaceae, Cryptococcus Public Health Service, Centers for Disease Control and Species: Cryptococcus gattii Prevention, and National Institutes of Health. Biosafety in Strain: C10 Microbiological and Biomedical Laboratories. 5th ed. Original Source: Cryptococcus gattii (C. gattii), strain C10 Washington, DC: U.S. Government Printing Office, 2009; see was isolated from an unknown human source in the Pacific www.cdc.gov/biosafety/publications/bmbl5/index.htm. Northwest region of North America.1 Comments: C. gattii, strain C10 was deposited as lineage Disclaimers: VGIIa and resistant to azoles.1 You are authorized to use this product for research use only. The Cryptococcus species complex is comprised of four It is not intended for human use. -

Post-Translational Regulation of Inflammasomes
OPEN Cellular & Molecular Immunology (2017) 14, 65–79 & 2017 CSI and USTC All rights reserved 2042-0226/17 www.nature.com/cmi REVIEW Post-translational regulation of inflammasomes Jie Yang1,2, Zhonghua Liu1 and Tsan Sam Xiao1 Inflammasomes play essential roles in immune protection against microbial infections. However, excessive inflammation is implicated in various human diseases, including autoinflammatory syndromes, diabetes, multiple sclerosis, cardiovascular disorders and neurodegenerative diseases. Therefore, precise regulation of inflammasome activities is critical for adequate immune protection while limiting collateral tissue damage. In this review, we focus on the emerging roles of post-translational modifications (PTMs) that regulate activation of the NLRP3, NLRP1, NLRC4, AIM2 and IFI16 inflammasomes. We anticipate that these types of PTMs will be identified in other types of and less well-characterized inflammasomes. Because these highly diverse and versatile PTMs shape distinct inflammatory responses in response to infections and tissue damage, targeting the enzymes involved in these PTMs will undoubtedly offer opportunities for precise modulation of inflammasome activities under various pathophysiological conditions. Cellular & Molecular Immunology (2017) 14, 65–79; doi:10.1038/cmi.2016.29; published online 27 June 2016 Keywords: inflammasome; phosphorylation; post-translational modifications; ubiquitination INTRODUCTION upstream sensor molecules through its PYD domain and The innate immune system relies on pattern recognition downstream -
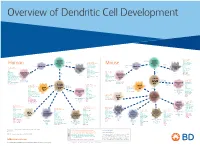
View Dendritic Cell Development Poster
Overview of Dendritic Cell Development Lineage–, CD45+, Common CD117 (c-kit) Common MHCII+, CD11c+ – + CD207 (Langerin) Myeloid CD117 (c-kit) Lineage , CD45 , Myeloid Progenitor MHCII (HLA-DR)+, CD11c+ Progenitor CD324 (E-Cadherin) Human Mouse CD326 (EpCAM) CD207 (Langerin) TGFb1 Cells CD11b, CD115 Cells CD14 Monocyte CD324 (E-Cadherin) Monocyte M-CSF CD11b – + Ly6C Langerhans CD24 Lineage , CD45 , M-CSF CD326 (EpCAM) MHCII (HLA-DR)+, CD11c+ Langerhans CD11blo Zbtb46– Cells CD172a (Sirp-α) CD16 CD1ahi, CD1c CD205 (DEC-205) Cells CSF F4/80 CD64 CD172a (Sirp-α) Lineage–, CD45+, FLT3L TLR3, TLR11 CD1a, CD1c Inflammatory CD369 (Dectin-1/CLEC7A) MHCII+, CD11c+ +/– CSF IL-15 CD8–, CD14– CD11b, CD14 CD371 (CLEC12A) CD64 Monocyte- FLT3L Inflammatory CD370 (Clec9a)– CD172a (Sirp-α) IL-15 CLEC6A CD11b derived lo Monocyte- CD206, CD209 (DC-SIGN) TLR1, TLR2, TLR3 , TLR6 CD209a (DC-SIGN) CD367 (DCIR/CLEC4A) DCs CD14– CD272 (BTLA)lo derived CD369 (Dectin-1/CLEC7A) DCs Common Ly-6C – + CD371 (CLEC12A) CD117 (c-kit) Lineage , CD45 , IL-1β, IL-6, IL-10, TLR1-6, TLR7-8, TLR10 Dendritic + lo CLEC6A – – CD135/FLT3 MHCII , CD11c IL-12, IL-23, TNF CD8a , CD14 IL-1β, IL-6 IL-10, Precursor TLR3lo, TLR4, TLR7, TLR8 CD45R (B220) IL-12, IL-23, TNF Plasmacytoid CD207 (Langerin)– Cells CD317 (BST-2) Common Lineage–, CD45+, FLT3L DCs Lineage–, CD45+, + Ly6C + lo/– CD207 IFN Type I + + Dendritic CD135/FLT3 MHCII (HLA-DR) , CD11c Lineage–, CD45+, IRF7, IRF8, BATF3hi Siglec-H MHCII (HLA-DR) , CD11c hi – + CD123 + + Dermal SpiB, Zbtb46 CD1a, CD64 CD1a Precursor CD117 (c-kit) -

Genetic Variant in 3' Untranslated Region of the Mouse Pycard Gene
bioRxiv preprint doi: https://doi.org/10.1101/2021.03.26.437184; this version posted March 26, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 2 3 Title: 4 Genetic Variant in 3’ Untranslated Region of the Mouse Pycard Gene Regulates Inflammasome 5 Activity 6 Running Title: 7 3’UTR SNP in Pycard regulates inflammasome activity 8 Authors: 9 Brian Ritchey1*, Qimin Hai1*, Juying Han1, John Barnard2, Jonathan D. Smith1,3 10 1Department of Cardiovascular & Metabolic Sciences, Lerner Research Institute, Cleveland Clinic, 11 Cleveland, OH 44195 12 2Department of Quantitative Health Sciences, Lerner Research Institute, Cleveland Clinic, Cleveland, OH 13 44195 14 3Department of Molecular Medicine, Cleveland Clinic Lerner College of Medicine of Case Western 15 Reserve University, Cleveland, OH 44195 16 *, These authors contributed equally to this study. 17 Address correspondence to Jonathan D. Smith: email [email protected]; ORCID ID 0000-0002-0415-386X; 18 mailing address: Cleveland Clinic, Box NC-10, 9500 Euclid Avenue, Cleveland, OH 44195, USA. 19 1 bioRxiv preprint doi: https://doi.org/10.1101/2021.03.26.437184; this version posted March 26, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 20 Abstract 21 Quantitative trait locus mapping for interleukin-1 release after inflammasome priming and activation 22 was performed on bone marrow-derived macrophages (BMDM) from an AKRxDBA/2 strain intercross. -

Cryptococcus Gattii Outbreak
The recent Cryptococcus gattii outbreak A deadly pathway… From Trees to Lungs to Brain ryptococcus gattii disease. The route of C has been in the infection is from breathing the news lately, due to a recent airborne organism, which outbreak in the Pacific becomes lodged in the Northwest. This new, more pulmonary tissues. virulent strain is expected to by Jay Hardy, CLS, SM (NRCM) spread further throughout the Symptoms Northwest and Northern California in the coming The pulmonary disease, Jay Hardy is the founder and months. known as cpryptococcosis, president of Hardy Diagnostics. develops slowly. After He began his career in exposure it can take two to microbiology as a Medical Technologist in Santa Barbara, twelve months for symptoms California. to appear, with the usual onset In 1980, he began at six or seven months. manufacturing culture media for the local hospitals. Today, The symptoms include: Hardy Diagnostics is the third largest media manufacturer in the U.S. • Cough that lasts weeks or months To ensure rapid and reliable turn around time, Hardy • Sharp chest pain Figure 1: An India ink stain Diagnostics maintains six • Unexplained shortness of showing the capsule surrounding distribution centers, and breath produces over 3,000 products the C. gattii cells in the yeast form used in clinical and industrial at 1,000X. Photo from Haley/CDC. • Severe headache microbiology laboratories • Confusion throughout the world. The organism is a fungus that • Fever is usually found in the soil • Night sweats www.HardyDiagnostics.com and on trees. It is not spread • Unintended weight loss from human to human or animal to human. -

ATP-Binding and Hydrolysis in Inflammasome Activation
molecules Review ATP-Binding and Hydrolysis in Inflammasome Activation Christina F. Sandall, Bjoern K. Ziehr and Justin A. MacDonald * Department of Biochemistry & Molecular Biology, Cumming School of Medicine, University of Calgary, 3280 Hospital Drive NW, Calgary, AB T2N 4Z6, Canada; [email protected] (C.F.S.); [email protected] (B.K.Z.) * Correspondence: [email protected]; Tel.: +1-403-210-8433 Academic Editor: Massimo Bertinaria Received: 15 September 2020; Accepted: 3 October 2020; Published: 7 October 2020 Abstract: The prototypical model for NOD-like receptor (NLR) inflammasome assembly includes nucleotide-dependent activation of the NLR downstream of pathogen- or danger-associated molecular pattern (PAMP or DAMP) recognition, followed by nucleation of hetero-oligomeric platforms that lie upstream of inflammatory responses associated with innate immunity. As members of the STAND ATPases, the NLRs are generally thought to share a similar model of ATP-dependent activation and effect. However, recent observations have challenged this paradigm to reveal novel and complex biochemical processes to discern NLRs from other STAND proteins. In this review, we highlight past findings that identify the regulatory importance of conserved ATP-binding and hydrolysis motifs within the nucleotide-binding NACHT domain of NLRs and explore recent breakthroughs that generate connections between NLR protein structure and function. Indeed, newly deposited NLR structures for NLRC4 and NLRP3 have provided unique perspectives on the ATP-dependency of inflammasome activation. Novel molecular dynamic simulations of NLRP3 examined the active site of ADP- and ATP-bound models. The findings support distinctions in nucleotide-binding domain topology with occupancy of ATP or ADP that are in turn disseminated on to the global protein structure. -

Idiopathic CD4 Lymphocytopenia Presenting As Refractory Cryptococcal Meningitis
Case Report Idiopathic CD4 lymphocytopenia presenting as refractory cryptococcal meningitis A. Sharma, V. Lal, M. Modi, D. Khurana, S. Bal, S. Prabhakar Department of Neurology, Postgraduate Institute of Medical Education and Research, Chandigarh-160 012, India Abstract Idiopathic CD4 T-lymphocytopenia (ICL) is a syndrome characterized by depletion of CD4 T-cells without evidence of human immunodeficiency virus (HIV) infection. There are a few reported cases of ICL associated with different diseases and clinical conditions, most commonly the opportunistic infections like Tuberculosis, fungal and parasitic diseases which are also seen in HIV-positive patients. We report a case without risk factors or laboratory evidence of HIV infection who presented with refractory cryptococcal meningitis and was found to have ICL. Key Words Cryptococcus, CD4, idiopathic For correspondence: Dr. Vivek Lal, Department of Neurology, PGIMER, Sector-12, Chandigarh-160 012, India. E-mail: [email protected] Ann Indian Acad Neurol 2010;13:136-8 [DOI: 10.4103/0972-2327.64646] Introduction and decreased pain and temperature sensation in the right half of the body, with sparing of the face. The neurological Idiopathic CD4 lymphocytopenia (ICL) was defi ned by the examination was otherwise normal. Plain magnetic resonance United States Centers for Disease Control and Prevention imaging (MRI) brain revealed a T2 hyperintensity in the (CDC) as a clinical condition in patients with depressed left thalamus, extending to involve the left corona radiata numbers of circulating CD4 T lymphocytes (<300 cells/µl or [Figure 1]. There was no meningeal enhancement on contrast- <20% of total T cells) at a minimum of two separate time points enhanced MRI. -

A Role for the Nlr Family Members Nlrc4 and Nlrp3 in Astrocytic Inflammasome Activation and Astrogliosis
A ROLE FOR THE NLR FAMILY MEMBERS NLRC4 AND NLRP3 IN ASTROCYTIC INFLAMMASOME ACTIVATION AND ASTROGLIOSIS Leslie C. Freeman A dissertation submitted to the faculty of the University of North Carolina at Chapel Hill in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Curriculum of Genetics and Molecular Biology. Chapel Hill 2016 Approved by: Jenny P. Y. Ting Glenn K. Matsushima Beverly H. Koller Silva S. Markovic-Plese Pauline. Kay Lund ©2016 Leslie C. Freeman ALL RIGHTS RESERVED ii ABSTRACT Leslie C. Freeman: A Role for the NLR Family Members NLRC4 and NLRP3 in Astrocytic Inflammasome Activation and Astrogliosis (Under the direction of Jenny P.Y. Ting) The inflammasome is implicated in many inflammatory diseases but has been primarily studied in the macrophage-myeloid lineage. Here we demonstrate a physiologic role for nucleotide-binding domain, leucine-rich repeat, CARD domain containing 4 (NLRC4) in brain astrocytes. NLRC4 has been primarily studied in the context of gram-negative bacteria, where it is required for the maturation of pro-caspase-1 to active caspase-1. We show the heightened expression of NLRC4 protein in astrocytes in a cuprizone model of neuroinflammation and demyelination as well as human multiple sclerotic brains. Similar to macrophages, NLRC4 in astrocytes is required for inflammasome activation by its known agonist, flagellin. However, NLRC4 in astrocytes also mediate inflammasome activation in response to lysophosphatidylcholine (LPC), an inflammatory molecule associated with neurologic disorders. In addition to NLRC4, astrocytic NLRP3 is required for inflammasome activation by LPC. Two biochemical assays show the interaction of NLRC4 with NLRP3, suggesting the possibility of a NLRC4-NLRP3 co-inflammasome.