Signalling Through C-Type Lectin Receptors
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse
Welcome to More Choice CD Marker Handbook For more information, please visit: Human bdbiosciences.com/eu/go/humancdmarkers Mouse bdbiosciences.com/eu/go/mousecdmarkers Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse CD3 CD3 CD (cluster of differentiation) molecules are cell surface markers T Cell CD4 CD4 useful for the identification and characterization of leukocytes. The CD CD8 CD8 nomenclature was developed and is maintained through the HLDA (Human Leukocyte Differentiation Antigens) workshop started in 1982. CD45R/B220 CD19 CD19 The goal is to provide standardization of monoclonal antibodies to B Cell CD20 CD22 (B cell activation marker) human antigens across laboratories. To characterize or “workshop” the antibodies, multiple laboratories carry out blind analyses of antibodies. These results independently validate antibody specificity. CD11c CD11c Dendritic Cell CD123 CD123 While the CD nomenclature has been developed for use with human antigens, it is applied to corresponding mouse antigens as well as antigens from other species. However, the mouse and other species NK Cell CD56 CD335 (NKp46) antibodies are not tested by HLDA. Human CD markers were reviewed by the HLDA. New CD markers Stem Cell/ CD34 CD34 were established at the HLDA9 meeting held in Barcelona in 2010. For Precursor hematopoetic stem cell only hematopoetic stem cell only additional information and CD markers please visit www.hcdm.org. Macrophage/ CD14 CD11b/ Mac-1 Monocyte CD33 Ly-71 (F4/80) CD66b Granulocyte CD66b Gr-1/Ly6G Ly6C CD41 CD41 CD61 (Integrin b3) CD61 Platelet CD9 CD62 CD62P (activated platelets) CD235a CD235a Erythrocyte Ter-119 CD146 MECA-32 CD106 CD146 Endothelial Cell CD31 CD62E (activated endothelial cells) Epithelial Cell CD236 CD326 (EPCAM1) For Research Use Only. -

Single Cell Transcriptome Atlas of Immune Cells in Human Small
bioRxiv preprint doi: https://doi.org/10.1101/721258; this version posted August 1, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Single cell transcriptome atlas of immune cells in human small 2 intestine and in celiac disease 3 4 Nader Atlasy1,a,4, Anna Bujko2,4, Peter B Brazda1,a, Eva Janssen-Megens1,a , Espen S. 5 Bækkevold2, Jørgen Jahnsen3, Frode L. Jahnsen2, Hendrik G. Stunnenberg1,a,* 6 7 8 1Department of Molecular Biology, Science Faculty, Radboud University, Nijmegen, The 9 Netherlands. 10 2 Department of Pathology, University of Oslo and Oslo University Hospital, Rikshospitalet, 11 Oslo, Norway 12 3 Department of Gastroenterology, Akershus University Hospital and University of Oslo, 13 Oslo, Norway. 14 15 acurrent address: Princess Maxima Centre for Pediatric Oncology, Heidelberglaan 25, 3584 16 CS Utrecht 17 18 19 4These authors contributed equally to this study 20 21 22 *Corresponding author: [email protected] 23 24 25 26 Celiac disease (CeD) is an autoimmune disorder in which ingestion of dietary gluten 27 triggers an immune reaction in the small intestine1,2. The CeD lesion is characterized by 28 crypt hyperplasia, villous atrophy and chronic inflammation with accumulation of 29 leukocytes both in the lamina propria (LP) and in the epithelium3, which eventually 30 leads to destruction of the intestinal epithelium1 and subsequent digestive complications 31 and higher risk of non-hodgkin lymphoma4. A lifetime gluten-free diet is currently the 32 only available treatment5. Gluten-specific LP CD4 T cells and cytotoxic intraepithelial 33 CD8+ T cells are thought to be central in disease pathology1,6-8, however, CeD is a 34 complex immune-mediated disorder and to date the findings are mostly based on 35 analysis of heterogeneous cell populations and on animal models. -

List of Genes Used in Cell Type Enrichment Analysis
List of genes used in cell type enrichment analysis Metagene Cell type Immunity ADAM28 Activated B cell Adaptive CD180 Activated B cell Adaptive CD79B Activated B cell Adaptive BLK Activated B cell Adaptive CD19 Activated B cell Adaptive MS4A1 Activated B cell Adaptive TNFRSF17 Activated B cell Adaptive IGHM Activated B cell Adaptive GNG7 Activated B cell Adaptive MICAL3 Activated B cell Adaptive SPIB Activated B cell Adaptive HLA-DOB Activated B cell Adaptive IGKC Activated B cell Adaptive PNOC Activated B cell Adaptive FCRL2 Activated B cell Adaptive BACH2 Activated B cell Adaptive CR2 Activated B cell Adaptive TCL1A Activated B cell Adaptive AKNA Activated B cell Adaptive ARHGAP25 Activated B cell Adaptive CCL21 Activated B cell Adaptive CD27 Activated B cell Adaptive CD38 Activated B cell Adaptive CLEC17A Activated B cell Adaptive CLEC9A Activated B cell Adaptive CLECL1 Activated B cell Adaptive AIM2 Activated CD4 T cell Adaptive BIRC3 Activated CD4 T cell Adaptive BRIP1 Activated CD4 T cell Adaptive CCL20 Activated CD4 T cell Adaptive CCL4 Activated CD4 T cell Adaptive CCL5 Activated CD4 T cell Adaptive CCNB1 Activated CD4 T cell Adaptive CCR7 Activated CD4 T cell Adaptive DUSP2 Activated CD4 T cell Adaptive ESCO2 Activated CD4 T cell Adaptive ETS1 Activated CD4 T cell Adaptive EXO1 Activated CD4 T cell Adaptive EXOC6 Activated CD4 T cell Adaptive IARS Activated CD4 T cell Adaptive ITK Activated CD4 T cell Adaptive KIF11 Activated CD4 T cell Adaptive KNTC1 Activated CD4 T cell Adaptive NUF2 Activated CD4 T cell Adaptive PRC1 Activated -

Identification of a Receptor Required for the Anti-Inflammatory Activity of IVIG
Identification of a receptor required for the INAUGURAL ARTICLE anti-inflammatory activity of IVIG Robert M. Anthonya, Fredrik Wermelinga,b, Mikael C. I. Karlssonb, and Jeffrey V. Ravetcha,1 aThe Laboratory of Molecular Genetics and Immunology, The Rockefeller University, New York, NY 10065; and bDepartment of Medicine, Karolinska Institutet, 171 76 Stockholm, Sweden This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on April 25, 2006. Contributed by Jeffrey V. Ravetch, October 11, 2008 (sent for review October 1, 2008) The anti-inflammatory activity of intravenous Ig (IVIG) results from patients suffering from autoimmune diseases (3). Monomeric IgG, a minor population of the pooled IgG molecules that contains purified from the serum of thousands of healthy donors (IVIG) is terminal ␣2,6-sialic acid linkages on their Fc-linked glycans. These a commonly administered at high doses (1–2 g/kg) for the treatment anti-inflammatory properties can be recapitulated with a fully of a number of autoimmune diseases, including immune-mediated recombinant preparation of appropriately sialylated IgG Fc frag- thrombocytopenia, chronic inflammatory demyelinating polyneu- ments. We now demonstrate that these sialylated Fcs require a ropathy, Kawasaki Disease and Guillain-Barre syndrome, and is specific C-type lectin, SIGN-R1, (specific ICAM-3 grabbing non- widely used in other autoimmune disorders (4–6). integrin-related 1) expressed on macrophages in the splenic mar- A number of hypotheses have been advanced to explain the ginal zone. Splenectomy, loss of SIGN-R1؉ cells in the splenic paradoxical activity of high dose IgG, and include models that marginal zone, blockade of the carbohydrate recognition domain attribute the activity to the polyclonal binding specificities, encoded (CRD) of SIGN-R1, or genetic deletion of SIGN-R1 abrogated the in the variable domains of the administered antibodies that may anti-inflammatory activity of IVIG or sialylated Fc fragments. -

Human Anogenital Monocyte-Derived Dendritic Cells and Langerin+Cdc2 Are Major HIV Target Cells
ARTICLE https://doi.org/10.1038/s41467-021-22375-x OPEN Human anogenital monocyte-derived dendritic cells and langerin+cDC2 are major HIV target cells Jake W. Rhodes 1,2,15, Rachel A. Botting 1,2,3,15, Kirstie M. Bertram1,2, Erica E. Vine 1,2, Hafsa Rana1,2, Heeva Baharlou1,2, Peter Vegh 3, Thomas R. O’Neil1,2, Anneliese S. Ashhurst4, James Fletcher3, Grant P. Parnell1,2, J. Dinny Graham1,2, Najla Nasr1,4, Jake J. K. Lim5, Laith Barnouti6, Peter Haertsch7, Martijn P. Gosselink 1,8, Angelina Di Re 1,8, Faizur Reza1,8, Grahame Ctercteko1,8, Gregory J. Jenkins9, Andrew J. Brooks10, Ellis Patrick 1,11, Scott N. Byrne1,4, Eric Hunter 12, Muzlifah A. Haniffa 3,13,14, ✉ Anthony L. Cunningham 1,2,16 & Andrew N. Harman 1,4,16 1234567890():,; Tissue mononuclear phagocytes (MNP) are specialised in pathogen detection and antigen presentation. As such they deliver HIV to its primary target cells; CD4 T cells. Most MNP HIV transmission studies have focused on epithelial MNPs. However, as mucosal trauma and inflammation are now known to be strongly associated with HIV transmission, here we examine the role of sub-epithelial MNPs which are present in a diverse array of subsets. We show that HIV can penetrate the epithelial surface to interact with sub-epithelial resident MNPs in anogenital explants and define the full array of subsets that are present in the human anogenital and colorectal tissues that HIV may encounter during sexual transmission. In doing so we identify two subsets that preferentially take up HIV, become infected and transmit the virus to CD4 T cells; CD14+CD1c+ monocyte-derived dendritic cells and langerin-expressing conventional dendritic cells 2 (cDC2). -

Microglial Function and Regulation During Development, Homeostasis and Alzheimer’S Disease
cells Review Microglial Function and Regulation during Development, Homeostasis and Alzheimer’s Disease Brad T. Casali and Erin G. Reed-Geaghan * Department of Pharmaceutical Sciences, Northeast Ohio Medical University, Rootstown, OH 44272, USA; [email protected] * Correspondence: [email protected] Abstract: Microglia are the resident immune cells of the brain, deriving from yolk sac progenitors that populate the brain parenchyma during development. During development and homeostasis, microglia play critical roles in synaptogenesis and synaptic plasticity, in addition to their primary role as immune sentinels. In aging and neurodegenerative diseases generally, and Alzheimer’s disease (AD) specifically, microglial function is altered in ways that significantly diverge from their homeostatic state, inducing a more detrimental inflammatory environment. In this review, we discuss the receptors, signaling, regulation and gene expression patterns of microglia that mediate their phenotype and function contributing to the inflammatory milieu of the AD brain, as well as strategies that target microglia to ameliorate the onset, progression and symptoms of AD. Keywords: microglia; inflammation; Alzheimer’s disease; neurodegenerative diseases; TREM2; neu- roinflammation Citation: Casali, B.T.; Reed-Geaghan, 1. Introduction E.G. Microglial Function and Microglia are the resident phagocytes of the central nervous system (CNS). In addition Regulation during Development, to their immunological role in maintaining CNS homeostasis, microglia play vital -

Macrophage Activation Markers, CD163 and CD206, in Acute-On-Chronic Liver Failure
cells Review Macrophage Activation Markers, CD163 and CD206, in Acute-on-Chronic Liver Failure Marlene Christina Nielsen 1 , Rasmus Hvidbjerg Gantzel 2 , Joan Clària 3,4 , Jonel Trebicka 3,5 , Holger Jon Møller 1 and Henning Grønbæk 2,* 1 Department of Clinical Biochemistry, Aarhus University Hospital, 8200 Aarhus N, Denmark; [email protected] (M.C.N.); [email protected] (H.J.M.) 2 Department of Hepatology & Gastroenterology, Aarhus University Hospital, 8200 Aarhus N, Denmark; [email protected] 3 European Foundation for the Study of Chronic Liver Failure (EF-CLIF), 08021 Barcelona, Spain; [email protected] (J.C.); [email protected] (J.T.) 4 Department of Biochemistry and Molecular Genetics, Hospital Clínic-IDIBAPS, 08036 Barcelona, Spain 5 Translational Hepatology, Department of Internal Medicine I, Goethe University Frankfurt, 60323 Frankfurt, Germany * Correspondence: [email protected]; Tel.: +45-21-67-92-81 Received: 1 April 2020; Accepted: 4 May 2020; Published: 9 May 2020 Abstract: Macrophages facilitate essential homeostatic functions e.g., endocytosis, phagocytosis, and signaling during inflammation, and express a variety of scavenger receptors including CD163 and CD206, which are upregulated in response to inflammation. In healthy individuals, soluble forms of CD163 and CD206 are constitutively shed from macrophages, however, during inflammation pathogen- and damage-associated stimuli induce this shedding. Activation of resident liver macrophages viz. Kupffer cells is part of the inflammatory cascade occurring in acute and chronic liver diseases. We here review the existing literature on sCD163 and sCD206 function and shedding, and potential as biomarkers in acute and chronic liver diseases with a particular focus on Acute-on-Chronic Liver Failure (ACLF). -

In Sickness and in Health: the Immunological Roles of the Lymphatic System
International Journal of Molecular Sciences Review In Sickness and in Health: The Immunological Roles of the Lymphatic System Louise A. Johnson MRC Human Immunology Unit, MRC Weatherall Institute of Molecular Medicine, University of Oxford, John Radcliffe Hospital, Headington, Oxford OX3 9DS, UK; [email protected] Abstract: The lymphatic system plays crucial roles in immunity far beyond those of simply providing conduits for leukocytes and antigens in lymph fluid. Endothelial cells within this vasculature are dis- tinct and highly specialized to perform roles based upon their location. Afferent lymphatic capillaries have unique intercellular junctions for efficient uptake of fluid and macromolecules, while expressing chemotactic and adhesion molecules that permit selective trafficking of specific immune cell subsets. Moreover, in response to events within peripheral tissue such as inflammation or infection, soluble factors from lymphatic endothelial cells exert “remote control” to modulate leukocyte migration across high endothelial venules from the blood to lymph nodes draining the tissue. These immune hubs are highly organized and perfectly arrayed to survey antigens from peripheral tissue while optimizing encounters between antigen-presenting cells and cognate lymphocytes. Furthermore, subsets of lymphatic endothelial cells exhibit differences in gene expression relating to specific func- tions and locality within the lymph node, facilitating both innate and acquired immune responses through antigen presentation, lymph node remodeling and regulation of leukocyte entry and exit. This review details the immune cell subsets in afferent and efferent lymph, and explores the mech- anisms by which endothelial cells of the lymphatic system regulate such trafficking, for immune surveillance and tolerance during steady-state conditions, and in response to infection, acute and Citation: Johnson, L.A. -

Single-Cell RNA Sequencing Demonstrates the Molecular and Cellular Reprogramming of Metastatic Lung Adenocarcinoma
ARTICLE https://doi.org/10.1038/s41467-020-16164-1 OPEN Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma Nayoung Kim 1,2,3,13, Hong Kwan Kim4,13, Kyungjong Lee 5,13, Yourae Hong 1,6, Jong Ho Cho4, Jung Won Choi7, Jung-Il Lee7, Yeon-Lim Suh8,BoMiKu9, Hye Hyeon Eum 1,2,3, Soyean Choi 1, Yoon-La Choi6,10,11, Je-Gun Joung1, Woong-Yang Park 1,2,6, Hyun Ae Jung12, Jong-Mu Sun12, Se-Hoon Lee12, ✉ ✉ Jin Seok Ahn12, Keunchil Park12, Myung-Ju Ahn 12 & Hae-Ock Lee 1,2,3,6 1234567890():,; Advanced metastatic cancer poses utmost clinical challenges and may present molecular and cellular features distinct from an early-stage cancer. Herein, we present single-cell tran- scriptome profiling of metastatic lung adenocarcinoma, the most prevalent histological lung cancer type diagnosed at stage IV in over 40% of all cases. From 208,506 cells populating the normal tissues or early to metastatic stage cancer in 44 patients, we identify a cancer cell subtype deviating from the normal differentiation trajectory and dominating the metastatic stage. In all stages, the stromal and immune cell dynamics reveal ontological and functional changes that create a pro-tumoral and immunosuppressive microenvironment. Normal resident myeloid cell populations are gradually replaced with monocyte-derived macrophages and dendritic cells, along with T-cell exhaustion. This extensive single-cell analysis enhances our understanding of molecular and cellular dynamics in metastatic lung cancer and reveals potential diagnostic and therapeutic targets in cancer-microenvironment interactions. 1 Samsung Genome Institute, Samsung Medical Center, Seoul 06351, Korea. -
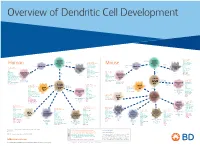
View Dendritic Cell Development Poster
Overview of Dendritic Cell Development Lineage–, CD45+, Common CD117 (c-kit) Common MHCII+, CD11c+ – + CD207 (Langerin) Myeloid CD117 (c-kit) Lineage , CD45 , Myeloid Progenitor MHCII (HLA-DR)+, CD11c+ Progenitor CD324 (E-Cadherin) Human Mouse CD326 (EpCAM) CD207 (Langerin) TGFb1 Cells CD11b, CD115 Cells CD14 Monocyte CD324 (E-Cadherin) Monocyte M-CSF CD11b – + Ly6C Langerhans CD24 Lineage , CD45 , M-CSF CD326 (EpCAM) MHCII (HLA-DR)+, CD11c+ Langerhans CD11blo Zbtb46– Cells CD172a (Sirp-α) CD16 CD1ahi, CD1c CD205 (DEC-205) Cells CSF F4/80 CD64 CD172a (Sirp-α) Lineage–, CD45+, FLT3L TLR3, TLR11 CD1a, CD1c Inflammatory CD369 (Dectin-1/CLEC7A) MHCII+, CD11c+ +/– CSF IL-15 CD8–, CD14– CD11b, CD14 CD371 (CLEC12A) CD64 Monocyte- FLT3L Inflammatory CD370 (Clec9a)– CD172a (Sirp-α) IL-15 CLEC6A CD11b derived lo Monocyte- CD206, CD209 (DC-SIGN) TLR1, TLR2, TLR3 , TLR6 CD209a (DC-SIGN) CD367 (DCIR/CLEC4A) DCs CD14– CD272 (BTLA)lo derived CD369 (Dectin-1/CLEC7A) DCs Common Ly-6C – + CD371 (CLEC12A) CD117 (c-kit) Lineage , CD45 , IL-1β, IL-6, IL-10, TLR1-6, TLR7-8, TLR10 Dendritic + lo CLEC6A – – CD135/FLT3 MHCII , CD11c IL-12, IL-23, TNF CD8a , CD14 IL-1β, IL-6 IL-10, Precursor TLR3lo, TLR4, TLR7, TLR8 CD45R (B220) IL-12, IL-23, TNF Plasmacytoid CD207 (Langerin)– Cells CD317 (BST-2) Common Lineage–, CD45+, FLT3L DCs Lineage–, CD45+, + Ly6C + lo/– CD207 IFN Type I + + Dendritic CD135/FLT3 MHCII (HLA-DR) , CD11c Lineage–, CD45+, IRF7, IRF8, BATF3hi Siglec-H MHCII (HLA-DR) , CD11c hi – + CD123 + + Dermal SpiB, Zbtb46 CD1a, CD64 CD1a Precursor CD117 (c-kit) -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Signal Integration of Syk-Coupled C-Type Lectin Receptors
Contact, Collaboration, and Conflict: Signal Integration of Syk-Coupled C-Type Lectin Receptors This information is current as Jenny Ostrop and Roland Lang of September 27, 2021. J Immunol 2017; 198:1403-1414; ; doi: 10.4049/jimmunol.1601665 http://www.jimmunol.org/content/198/4/1403 Downloaded from References This article cites 202 articles, 75 of which you can access for free at: http://www.jimmunol.org/content/198/4/1403.full#ref-list-1 Why The JI? Submit online. http://www.jimmunol.org/ • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average by guest on September 27, 2021 Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2017 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. Th eJournal of Brief Reviews Immunology Contact, Collaboration, and Conflict: Signal Integration of Syk-Coupled C-Type Lectin Receptors Jenny Ostrop*,† and Roland Lang‡ Several spleen tyrosine kinase–coupled C-type lectin of TLRs and crosstalk of TLR signaling have been studied for receptors (CLRs) have emerged as important pattern almost two decades.