Looking for Therapeutic Antibodies in Next Generation Sequencing Repositories
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Recent Advances in Inflammatory Bowel Disease: Mucosal Immune
Recent advances in basic science Recent advances in inflammatory bowel disease: Gut: first published as 10.1136/gutjnl-2012-303955 on 8 October 2013. Downloaded from mucosal immune cells in intestinal inflammation M Zaeem Cader,1,2 Arthur Kaser1 1Department of Medicine, ABSTRACT Recent years have seen a rapid and exciting Division of Gastroenterology & The intestine and its immune system have evolved to expansion in our understanding of the mucosal Hepatology, University of Cambridge, Addenbrooke’s meet the extraordinary task of maintaining tolerance to immune system with novel insights into environ- Hospital, Cambridge, UK the largest, most complex and diverse microbial mental influences of diet and the microbiota; the 2Wellcome Trust PhD commensal habitat, while meticulously attacking and convergence and integration of fundamental cellu- Programme for Clinicians, containing even minute numbers of occasionally lar processes such as autophagy, microbial sensing Cambridge Institute for incoming pathogens. While our understanding is still far and endoplasmic reticulum (ER) stress; as well as Medical Research, School of Clinical Medicine, University of from complete, recent studies have provided exciting the discovery of new cell types, for example innate Cambridge, Cambridge, UK novel insights into the complex interplay of the many lymphoid cells (ILCs). distinct intestinal immune cell types as well as the The gut is unlike the systemic immune system in Correspondence to discovery of entirely new cell subsets. These studies have several respects and much of the extensive reper- Dr Arthur Kaser, Department of Medicine, Division of also revealed how proper development and function of toire of immune cells and their characteristics are Gastroenterology and the intestinal immune system is dependent on its specific indeed unique to the intestine. -

Predictive QSAR Tools to Aid in Early Process Development of Monoclonal Antibodies
Predictive QSAR tools to aid in early process development of monoclonal antibodies John Micael Andreas Karlberg Published work submitted to Newcastle University for the degree of Doctor of Philosophy in the School of Engineering November 2019 Abstract Monoclonal antibodies (mAbs) have become one of the fastest growing markets for diagnostic and therapeutic treatments over the last 30 years with a global sales revenue around $89 billion reported in 2017. A popular framework widely used in pharmaceutical industries for designing manufacturing processes for mAbs is Quality by Design (QbD) due to providing a structured and systematic approach in investigation and screening process parameters that might influence the product quality. However, due to the large number of product quality attributes (CQAs) and process parameters that exist in an mAb process platform, extensive investigation is needed to characterise their impact on the product quality which makes the process development costly and time consuming. There is thus an urgent need for methods and tools that can be used for early risk-based selection of critical product properties and process factors to reduce the number of potential factors that have to be investigated, thereby aiding in speeding up the process development and reduce costs. In this study, a framework for predictive model development based on Quantitative Structure- Activity Relationship (QSAR) modelling was developed to link structural features and properties of mAbs to Hydrophobic Interaction Chromatography (HIC) retention times and expressed mAb yield from HEK cells. Model development was based on a structured approach for incremental model refinement and evaluation that aided in increasing model performance until becoming acceptable in accordance to the OECD guidelines for QSAR models. -
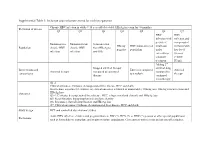
Inclusion and Exclusion Criteria for Each Key Question
Supplemental Table 1: Inclusion and exclusion criteria for each key question Chronic HBV infection in adults ≥ 18 year old (detectable HBsAg in serum for >6 months) Definition of disease Q1 Q2 Q3 Q4 Q5 Q6 Q7 HBV HBV infection with infection and persistent compensated Immunoactive Immunotolerant Seroconverted HBeAg HBV mono-infected viral load cirrhosis with Population chronic HBV chronic HBV from HBeAg to negative population under low level infection infection anti-HBe entecavir or viremia tenofovir (<2000 treatment IU/ml) Adding 2nd Stopped antiviral therapy antiviral drug Interventions and Entecavir compared Antiviral Antiviral therapy compared to continued compared to comparisons to tenofovir therapy therapy continued monotherapy Q1-2: Clinical outcomes: Cirrhosis, decompensated liver disease, HCC and death Intermediate outcomes (if evidence on clinical outcomes is limited or unavailable): HBsAg loss, HBeAg seroconversion and Outcomes HBeAg loss Q3-4: Cirrhosis, decompensated liver disease, HCC, relapse (viral and clinical) and HBsAg loss Q5: Renal function, hypophosphatemia and bone density Q6: Resistance, flare/decompensation and HBeAg loss Q7: Clinical outcomes: Cirrhosis, decompensated liver disease, HCC and death Study design RCT and controlled observational studies Acute HBV infection, children and pregnant women, HIV (+), HCV (+) or HDV (+) persons or other special populations Exclusions such as hemodialysis, transplant, and treatment failure populations. Co treatment with steroids and uncontrolled studies. Supplemental Table 2: Detailed Search Strategy: Ovid Database(s): Embase 1988 to 2014 Week 37, Ovid MEDLINE(R) In-Process & Other Non- Indexed Citations and Ovid MEDLINE(R) 1946 to Present, EBM Reviews - Cochrane Central Register of Controlled Trials August 2014, EBM Reviews - Cochrane Database of Systematic Reviews 2005 to July 2014 Search Strategy: # Searches Results 1 exp Hepatitis B/dt 26410 ("hepatitis B" or "serum hepatitis" or "hippie hepatitis" or "injection hepatitis" or 2 178548 "hepatitis type B").mp. -

(12) United States Patent (10) Patent No.: US 9.468,689 B2 Zeng Et Al
USOO9468689B2 (12) United States Patent (10) Patent No.: US 9.468,689 B2 Zeng et al. (45) Date of Patent: *Oct. 18, 2016 (54) ULTRAFILTRATION CONCENTRATION OF (56) References Cited ALLOTYPE SELECTED ANTIBODES FOR SMALL-VOLUME ADMINISTRATION U.S. PATENT DOCUMENTS 5,429,746 A 7/1995 Shadle et al. (71) Applicant: Immunomedics, Inc., Morris Plains, NJ 5,789,554 A 8/1998 Leung et al. (US) 6,171,586 B1 1/2001 Lam et al. 6,187,287 B1 2/2001 Leung et al. (72) Inventors: Li Zeng, Edison, NJ (US); Rohini 6,252,055 B1 6/2001 Relton Mitra, Brigdewater, NJ (US); Edmund 6,676,924 B2 1/2004 Hansen et al. 6,870,034 B2 3/2005 Breece et al. A. Rossi, Woodland Park, NJ (US); 6,893,639 B2 5/2005 Levy et al. Hans J. Hansen, Picayune, MS (US); 6,991,790 B1 1/2006 Lam et al. David M. Goldenberg, Mendham, NJ 7,038,017 B2 5, 2006 Rinderknecht et al. (US) 7,074,403 B1 7/2006 Goldenberg et al. 7,109,304 B2 9, 2006 Hansen et al. 7,138,496 B2 11/2006 Hua et al. (73) Assignee: Immunomedics, Inc., Morris Plains, NJ 7,151,164 B2 * 12/2006 Hansen et al. ............. 530,387.3 (US) 7,238,785 B2 7/2007 Govindan et al. 7,251,164 B2 7/2007 Okhonin et al. (*) Notice: Subject to any disclaimer, the term of this 7.282,567 B2 10/2007 Goldenberg et al. patent is extended or adjusted under 35 7,300,655 B2 11/2007 Hansen et al. -

Activating Death Receptor DR5 As a Therapeutic Strategy for Rhabdomyosarcoma
International Scholarly Research Network ISRN Oncology Volume 2012, Article ID 395952, 10 pages doi:10.5402/2012/395952 Review Article Activating Death Receptor DR5 as a Therapeutic Strategy for Rhabdomyosarcoma Zhigang Kang,1, 2 Shi-Yong Sun,3 and Liang Cao1 1 Genetics Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD 20892, USA 2 Laboratory of Proteomics and Analytical Technologies, SAIC-Frederick, Inc., NCI Frederick, Frederick, MD 21702, USA 3 Department of Hematology and Medical Oncology, Winship Cancer Institute, Emory University School of Medicine, Atlanta, GA 30322, USA Correspondence should be addressed to Liang Cao, [email protected] Received 4 January 2012; Accepted 24 January 2012 Academic Editors: E. Boven and S. Mandruzzato Copyright © 2012 Zhigang Kang et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Rhabdomyosarcoma (RMS) is the most common soft tissue sarcoma in children. It is believed to arise from skeletal muscle progenitors, preserving the expression of genes critical for embryonic myogenic development such as MYOD1 and myogenin. RMS is classified as embryonal, which is more common in younger children, or alveolar, which is more prevalent in elder children and adults. Despite aggressive management including surgery, radiation, and chemotherapy, the outcome for children with metastatic RMS is dismal, and the prognosis has remained unchanged for decades. Apoptosis is a highly regulated process critical for embryonic development and tissue and organ homeostasis. Like other types of cancers, RMS develops by evading intrinsic apoptosis via mutations in the p53 tumor suppressor gene. -

Comparative Safety and Efficacy of Anti-PD-1 Monotherapy, Chemotherapy
Lv et al. Journal for ImmunoTherapy of Cancer (2019) 7:159 https://doi.org/10.1186/s40425-019-0636-7 COMMENTARY Open Access Comparative safety and efficacy of anti-PD- 1 monotherapy, chemotherapy alone, and their combination therapy in advanced nasopharyngeal carcinoma: findings from recent advances in landmark trials Jia-Wei Lv1†, Jun-Yan Li1†, Lin-Na Luo2†, Zi-Xian Wang2* and Yu-Pei Chen1* Abstract Recent phase 1–2 trials reported manageable safety profiles and promising antitumor activities of anti-PD-1 drugs (pembrolizumab, nivolumab, camrelizumab and JS001) with/without chemotherapy in recurrent/metastatic nasopharyngeal carcinoma (RM-NPC), however head-to-head comparison among these regimens is lacking. We aimed to comprehensively compare the efficacy and safety of different anti-PD-1 drugs, standard chemotherapy, and their combination therapy in RM-NPC. Adverse event (AE) and objective response rate (ORR) were assessed. The pooled incidence rates of grade 1–5/3–5 AEs were 74.1%/29.6, 54.2%/17.4, 92.3%/24.5, 96.8%/16.1, 91.2%/42.8, and 100%/87.9% for pembrolizumab, nivolumab, JS001, camrelizumab, chemotherapy and camrelizumab+chemotherapy, respectively, which suggested that nivolumab and pembrolizumab exhibited the optimal safety regarding grade 1–5 AEs whereas camrelizumab and nivolumab regarding grade 3–5 AEs. As second- or later-line therapy, ORR was higher with camrelizumab (34.1%), followed by pembrolizumab (26.3%), JS001 (23.3%), and nivolumab (19.0%); whereas ORR with first-line nivolumab reached 40%. Additionally, first-line camrelizumab+chemotherapy achieved a dramatically higher ORR than that with chemotherapy alone (90.9% vs. -

WO 2015/028850 Al 5 March 2015 (05.03.2015) P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2015/028850 Al 5 March 2015 (05.03.2015) P O P C T (51) International Patent Classification: AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, C07D 519/00 (2006.01) A61P 39/00 (2006.01) BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, C07D 487/04 (2006.01) A61P 35/00 (2006.01) DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, A61K 31/5517 (2006.01) A61P 37/00 (2006.01) HN, HR, HU, ID, IL, IN, IS, JP, KE, KG, KN, KP, KR, A61K 47/48 (2006.01) KZ, LA, LC, LK, LR, LS, LT, LU, LY, MA, MD, ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, (21) International Application Number: OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, PCT/IB2013/058229 SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, (22) International Filing Date: TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, 2 September 2013 (02.09.2013) ZW. (25) Filing Language: English (84) Designated States (unless otherwise indicated, for every kind of regional protection available): ARIPO (BW, GH, (26) Publication Language: English GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, SZ, TZ, (71) Applicant: HANGZHOU DAC BIOTECH CO., LTD UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, TJ, [US/CN]; Room B2001-B2019, Building 2, No 452 Sixth TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, Street, Hangzhou Economy Development Area, Hangzhou EE, ES, FI, FR, GB, GR, HR, HU, IE, IS, IT, LT, LU, LV, City, Zhejiang 310018 (CN). -

Classification Decisions Taken by the Harmonized System Committee from the 47Th to 60Th Sessions (2011
CLASSIFICATION DECISIONS TAKEN BY THE HARMONIZED SYSTEM COMMITTEE FROM THE 47TH TO 60TH SESSIONS (2011 - 2018) WORLD CUSTOMS ORGANIZATION Rue du Marché 30 B-1210 Brussels Belgium November 2011 Copyright © 2011 World Customs Organization. All rights reserved. Requests and inquiries concerning translation, reproduction and adaptation rights should be addressed to [email protected]. D/2011/0448/25 The following list contains the classification decisions (other than those subject to a reservation) taken by the Harmonized System Committee ( 47th Session – March 2011) on specific products, together with their related Harmonized System code numbers and, in certain cases, the classification rationale. Advice Parties seeking to import or export merchandise covered by a decision are advised to verify the implementation of the decision by the importing or exporting country, as the case may be. HS codes Classification No Product description Classification considered rationale 1. Preparation, in the form of a powder, consisting of 92 % sugar, 6 % 2106.90 GRIs 1 and 6 black currant powder, anticaking agent, citric acid and black currant flavouring, put up for retail sale in 32-gram sachets, intended to be consumed as a beverage after mixing with hot water. 2. Vanutide cridificar (INN List 100). 3002.20 3. Certain INN products. Chapters 28, 29 (See “INN List 101” at the end of this publication.) and 30 4. Certain INN products. Chapters 13, 29 (See “INN List 102” at the end of this publication.) and 30 5. Certain INN products. Chapters 28, 29, (See “INN List 103” at the end of this publication.) 30, 35 and 39 6. Re-classification of INN products. -

Tanibirumab (CUI C3490677) Add to Cart
5/17/2018 NCI Metathesaurus Contains Exact Match Begins With Name Code Property Relationship Source ALL Advanced Search NCIm Version: 201706 Version 2.8 (using LexEVS 6.5) Home | NCIt Hierarchy | Sources | Help Suggest changes to this concept Tanibirumab (CUI C3490677) Add to Cart Table of Contents Terms & Properties Synonym Details Relationships By Source Terms & Properties Concept Unique Identifier (CUI): C3490677 NCI Thesaurus Code: C102877 (see NCI Thesaurus info) Semantic Type: Immunologic Factor Semantic Type: Amino Acid, Peptide, or Protein Semantic Type: Pharmacologic Substance NCIt Definition: A fully human monoclonal antibody targeting the vascular endothelial growth factor receptor 2 (VEGFR2), with potential antiangiogenic activity. Upon administration, tanibirumab specifically binds to VEGFR2, thereby preventing the binding of its ligand VEGF. This may result in the inhibition of tumor angiogenesis and a decrease in tumor nutrient supply. VEGFR2 is a pro-angiogenic growth factor receptor tyrosine kinase expressed by endothelial cells, while VEGF is overexpressed in many tumors and is correlated to tumor progression. PDQ Definition: A fully human monoclonal antibody targeting the vascular endothelial growth factor receptor 2 (VEGFR2), with potential antiangiogenic activity. Upon administration, tanibirumab specifically binds to VEGFR2, thereby preventing the binding of its ligand VEGF. This may result in the inhibition of tumor angiogenesis and a decrease in tumor nutrient supply. VEGFR2 is a pro-angiogenic growth factor receptor -

(12) United States Patent (10) Patent No.: US 9,161,992 B2 Jefferies Et Al
US009 161992B2 (12) United States Patent (10) Patent No.: US 9,161,992 B2 Jefferies et al. (45) Date of Patent: Oct. 20, 2015 (54) P97 FRAGMENTS WITH TRANSFER 4,683.202 A 7, 1987 Mullis ACTIVITY 4,704,362 A 11/1987 Itakura et al. 4,766,075 A 8, 1988 Goeddeletal. (71) Applicant: biosis Technologies, Inc., Richmond 4,800,1594,784.950 A 11/19881/1989 MullisHagen et al. (CA) 4,801,542 A 1/1989 Murray et al. 4.866,042 A 9, 1989 Neuwelt (72) Inventors: Wilfred Jefferies, South Surrey (CA); 4,935,349 A 6/1990 McKnight et al. Mei Mei Tian, Coquitlam (CA): 4.946,778 A 8, 1990 Ladner et al. Timothy Vitalis, Vancouver (CA) 5,091,513 A 2f1992 Huston et al. 5,132,405 A 7, 1992 Huston et al. (73) Assignee: biOasis Technologies, Inc., British 5, 186,941 A 2f1993 Callahan et al. Columbia (CA) 5,672,683 A 9, 1997 Friden et al. 5,677,171 A 10, 1997 Hudziak et al. c - r 5,720,937 A 2f1998 Hudziak et al. (*) Notice: Subject to any disclaimer, the term of this 5,720,954. A 2f1998 Hudziak et al. patent is extended or adjusted under 35 5,725,856 A 3, 1998 Hudziak et al. U.S.C. 154(b) by 0 days. 5,770,195 A 6/1998 Hudziak et al. 5,772,997 A 6/1998 Hudziak et al. (21) Appl. No.: 14/226,506 5,844,093 A 12/1998 Kettleborough et al. 5,962,012 A 10, 1999 Lin et al. -

Download PDF Document
Journal for ImmunoTherapy of Cancer 2019, 7(Suppl 1):283 https://doi.org/10.1186/s40425-019-0764-0 MEETINGABSTRACTS Open Access 34th Annual Meeting & Pre-Conference Programs of the Society for Immunotherapy of Cancer (SITC 2019): part 2 National Harbor, MD, USA. 10 November 2019 Published: 6 November 2019 About this supplement These abstracts have been published as part of Journal for ImmunoTherapy of Cancer Volume 7 Supplement 1, 2019. The full contents of the supplement are available online at https://jitc.biomedcentral.com/articles/supplements/volume-7-supplement-1. Please note that this is part 2 of 2. Poster Presentations median survival; 3) FTN203 did not negate the efficacy of anti- PD-1 or anti-PD-L1 immunotherapy antibody when combined; 4) Biomarkers, Immune Monitoring, and Novel FTN203 significantly improved the efficacy of anti-PD-1 by further Technologies reducing the tumor growth (by 80% on day 26, P=0.046) and in- creasing the median survival (by 5 days or 14%, Log-rank test P= P501 0.031), against the combo of COMPLETE and anti-PD-1; 5) None Dietary deprivation of non-essential amino acids improves anti-PD- of the mono or combo treatments caused body weight loss. 1 immunotherapy in murine colon cancer Conclusions Zehui Li, PhD, Grace Yang, PhD, Shuang Zhou, PhD, Xin Wang, MD, PhD, Our data supports the use of dietary NEAA deprivation to improve Xiyan Li, PhD the efficacy of anti-PD-1 or anti-PD-L1 immunotherapy for colorectal Filtricine, Inc., Santa Clara, CA, United States cancer without noticeable side effects. -

PD-1 / PD-L1 Combination Therapies
PD-1 / PD-L1 Combination Therapies Jacob Plieth & Edwin Elmhirst – May 2017 Foreword Sprinkling the immuno-oncology dust When in November 2015 EP Vantage published its first immuno- oncology analysis we identified 215 studies of anti-PD-1/PD-L1 projects combined with other approaches, and called this an important industry theme. It is a measure of how central combos have become that today, barely 18 months on, that total has been blown out of the water. No fewer than 765 studies involving combinations of PD-1 or PD-L1 assets are now listed on the Clinicaltrials.gov registry. This dazzling array owes much to the transformational nature of the data seen with the first wave of anti-PD-1/PD-L1 MAbs. It also indicates how central combinations will be in extending immuno-oncology beyond just a handful of cancers, and beyond certain patient subgroups. But the combo effort is as much about extending the reach of currently available drugs like Keytruda, Opdivo and Tecentriq as it is about making novel approaches viable by combining them with PD-1/PD-L1. On a standalone basis several of the industry’s novel oncology projects have underwhelmed. As data are generated it will therefore be vital for investors to tease out the effect of combinations beyond that of monotherapy, and it is doubtful whether the sprinkling of magic immuno-oncology dust will come to the rescue of substandard products. This is not stopping many companies, as the hundreds of combination studies identified here show. This report aims to quantify how many trials are ongoing with which assets and in which cancer indications, as well as suggesting reasons why some of the most popular approaches are being pursued.