Supplementary Tables and Figures
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Evolution and Diversification of the Plant Gibberellin Receptor GID1
Evolution and diversification of the plant gibberellin receptor GID1 Hideki Yoshidaa,b, Eiichi Tanimotoc, Takaaki Hiraia, Yohei Miyanoirid,e, Rie Mitania, Mayuko Kawamuraa, Mitsuhiro Takedad,f, Sayaka Takeharaa, Ko Hiranoa, Masatsune Kainoshod,g, Takashi Akagih, Makoto Matsuokaa,1, and Miyako Ueguchi-Tanakaa,1 aBioscience and Biotechnology Center, Nagoya University, Nagoya, 464-8601 Aichi, Japan; bKihara Institute for Biological Research, Yokohama City University, Yokohama, 244-0813 Kanagawa, Japan; cGraduate School of Natural Sciences, Nagoya City University, Nagoya, 467-8501 Aichi, Japan; dStructural Biology Research Center, Graduate School of Science, Nagoya University, Nagoya, 464-8601 Aichi, Japan; eResearch Center for State-of-the-Art Functional Protein Analysis, Institute for Protein Research, Osaka University, Suita, 565-0871 Osaka, Japan; fDepartment of Structural BioImaging, Faculty of Life Sciences, Kumamoto University, 862-0973 Kumamoto, Japan; gGraduate School of Science and Engineering, Tokyo Metropolitan University, Hachioji, 192-0397 Tokyo, Japan; and hGraduate School of Agriculture, Kyoto University, 606-8502 Kyoto, Japan Edited by Mark Estelle, University of California, San Diego, La Jolla, CA, and approved July 10, 2018 (received for review April 9, 2018) The plant gibberellin (GA) receptor GID1 shows sequence similarity erwort Marchantia polymorpha (5–7). Furthermore, Hirano et al. to carboxylesterase (CXE). Here, we report the molecular evolution (5) reported that GID1s in the lycophyte Selaginella moellen- of GID1 from establishment to functionally diverse forms in dorffii (SmGID1s) have unique properties in comparison with eudicots. By introducing 18 mutagenized rice GID1s into a rice angiosperm GID1s: namely, lower affinity to bioactive GAs and gid1 null mutant, we identified the amino acids crucial for higher affinity to inactive GAs (lower specificity). -
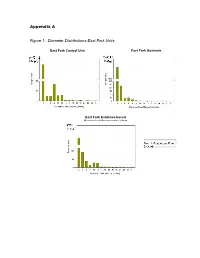
Appendix a Figure 1: Diameter Distributions-East Fork Units
Appendix A Figure 1: Diameter Distributions-East Fork Units East Fork Control Unit East Fork Goshawk Guidelines Unit East Fork Evidence-based Ecological Restoration Unit Figure 2: Diameter Distributions-Redondo Units Redondo Control Unit Redondo Goshawk Guidelines Unit Redondo Evidence-based Ecological Restoration Unit Figure 3: FlamMap Simulated Crown Fire Behavior: East Fork Project Area 20 mph Southwest Wind 25 mph 30 mph 35 mph 40 mph Map Legend Crown Fire Activity N No crown fire Passive crown activity fire (torching) Active crown fire (spreading) Important Note: Expected Crown Fire Behavior by Percent The 97th percentile of low fuel of Project Area moistures for June from 34 No Crown Passive years of data from the Kaibab Fire Crown Fire Active National Forest, Arizona were used in all simulations. This Windspeed Activity (torching) Crown Fire 20 79.9 0.9 19.2 input represents potential 25 71.2 1.9 26.9 extremes of fire weather during the peak of the fire 30 64.9 2.9 32.3 season, not average fire 35 57.3 4.7 38.0 season conditions. 40 48.7 9.4 42.0 Figure 4: FlamMap Simulated Crown Fire Behavior: Redondo Project Area 20 mph Southwest Wind 25 mph 30 mph 35 mph 40 mph Map Legend Crown Fire Activity N Highway 4 Valles Caldera No crown fire Passive crown activity entrance road fire (torching) Active crown fire (spreading) Important Note: Expected Crown Fire Behavior by Percent The 97th percentile of low fuel of Project Area moistures for June from 34 Passive years of data from the Kaibab No Crown Crown Fire Active Crown National Forest, Arizona were Windspeed Fire Activity (torching) Fire used in all simulations. -

Washington Flora Checklist a Checklist of the Vascular Plants of Washington State Hosted by the University of Washington Herbarium
Washington Flora Checklist A checklist of the Vascular Plants of Washington State Hosted by the University of Washington Herbarium The Washington Flora Checklist aims to be a complete list of the native and naturalized vascular plants of Washington State, with current classifications, nomenclature and synonymy. The checklist currently contains 3,929 terminal taxa (species, subspecies, and varieties). Taxa included in the checklist: * Native taxa whether extant, extirpated, or extinct. * Exotic taxa that are naturalized, escaped from cultivation, or persisting wild. * Waifs (e.g., ballast plants, escaped crop plants) and other scarcely collected exotics. * Interspecific hybrids that are frequent or self-maintaining. * Some unnamed taxa in the process of being described. Family classifications follow APG IV for angiosperms, PPG I (J. Syst. Evol. 54:563?603. 2016.) for pteridophytes, and Christenhusz et al. (Phytotaxa 19:55?70. 2011.) for gymnosperms, with a few exceptions. Nomenclature and synonymy at the rank of genus and below follows the 2nd Edition of the Flora of the Pacific Northwest except where superceded by new information. Accepted names are indicated with blue font; synonyms with black font. Native species and infraspecies are marked with boldface font. Please note: This is a working checklist, continuously updated. Use it at your discretion. Created from the Washington Flora Checklist Database on September 17th, 2018 at 9:47pm PST. Available online at http://biology.burke.washington.edu/waflora/checklist.php Comments and questions should be addressed to the checklist administrators: David Giblin ([email protected]) Peter Zika ([email protected]) Suggested citation: Weinmann, F., P.F. Zika, D.E. Giblin, B. -

A Vascular Flora Inventory
A Vascular Flora Inventory Ottawa Sands Ottawa County Parks, Michigan September 2020 Prepared by William Martinus & Associates Financial assistance for this project was provided, in part, by the Coastal Management Program, Water Resources Division, Michigan Department of Environment, Great Lakes, and Energy, under the National Coastal Zone Management program, through a grant from the National Oceanic and Atmospheric Administration, U.S. Department of Commerce. The statements, findings, conclusions, and recommendations in this report are those of the Ottawa County Parks & Recreation Commission and do not necessarily reflect the views of the Michigan Department of Environment, Great Lakes, and Energy or the National Oceanic and Atmospheric Administration. 1 Table of Contents I. Introduction and Purpose 3 II. Overview 3 III. Plant Communities 4 IV. Endangered, Threatened, and Special Concern Species 5 V. Species Lists 7 VI. References 21 2 I. Introduction and Purpose Ottawa Sands, Ottawa County Parks, consists of 345 acres including an 80-acre inland lake, natural forests, coastal dunes, intermittent wetlands, inundated shrub swamp, and riparian marsh, shrub, and swamp communities. The eleven natural plant communities occurring on the site are listed along with hundreds of associated plant and animal species. - Ottawa Sands is located near the mouth of the Grand River in sections 17, 18 and 20 of Spring Lake Township, Ottawa County, in Western Michigan. - Property includes 5,585 feet of Grand River frontage. - A Floristic Quality Assessment demonstrates that a diverse and extremely high-quality plant component exists at Ottawa Sands. Purpose - To gain an understanding of the vegetative plant communities and flora of western Ottawa County and central west Michigan area. -

Karyotype Evolution in Apomictic Boechera and the Origin of the Aberrant Chromosomes
The Plant Journal (2015) 82, 785–793 doi: 10.1111/tpj.12849 Karyotype evolution in apomictic Boechera and the origin of the aberrant chromosomes Terezie Mandakov a1, M. Eric Schranz2, Timothy F. Sharbel3, Hans de Jong4 and Martin A. Lysak1,* 1CEITEC – Central European Institute of Technology, Masaryk University, Brno, CZ-62500 Czech Republic, 2Plant Systematics Group, Wageningen University (WU), Droevendaalsesteeg 1, Wageningen, 6708 PB The Netherlands, 3Apomixis Research Group, Leibniz Institute of Plant Genetics and Crop Plant Research (IPK), Gatersleben, D-06466 Germany, and 4Laboratory of Genetics, Wageningen UR PSG, P.O. Box 16, Wageningen, 6700 AA The Netherlands Received 17 February 2015; revised 24 March 2015; accepted 1 April 2015; published online 10 April 2015. *For correspondence (e-mail [email protected]). SUMMARY Chromosome rearrangements may result in both decrease and increase of chromosome numbers. Here we have used comparative chromosome painting (CCP) to reconstruct the pathways of descending and ascend- ing dysploidy in the genus Boechera (tribe Boechereae, Brassicaceae). We describe the origin and structure of three Boechera genomes and establish the origin of the previously described aberrant Het and Del chro- mosomes found in Boechera apomicts with euploid (2n = 14) and aneuploid (2n = 15) chromosome number. CCP analysis allowed us to reconstruct the origin of seven chromosomes in sexual B. stricta and apomictic B. divaricarpa from the ancestral karyotype (n = 8) of Brassicaceae lineage I. Whereas three chromosomes (BS4, BS6, and BS7) retained their ancestral structure, five chromosomes were reshuffled by reciprocal translocations to form chromosomes BS1-BS3 and BS5. The reduction of the chromosome number (from x = 8tox= 7) was accomplished through the inactivation of a paleocentromere on chromosome BS5. -

100 Years of Change in the Flora of the Carolinas
ASTERACEAE 224 Zinnia Linnaeus 1759 (Zinnia) A genus of about 17 species, herbs, of sw. North America south to South America. References: Smith in FNA (2006c); Cronquist (1980)=SE. 1 Achenes wingless; receptacular bracts (chaff) toothed or erose on the lip..............................................................Z. peruviana 1 Achenes winged; receptacular bracts (chaff) with a differentiated fimbriate lip........................................................Z. violacea * Zinnia peruviana (Linnaeus) Linnaeus, Zinnia. Cp (GA, NC, SC): disturbed areas; rare (commonly cultivated), introduced from the New World tropics. May-November. [= FNA, K, SE; ? Z. pauciflora Linnaeus – S] * Zinnia violacea Cavanilles, Garden Zinnia. Cp (GA, NC, SC): disturbed areas; rare (commonly cultivated), introduced from the New World tropics. May-November. [= FNA, K; ? Z. elegans Jacquin – S, SE] BALSAMINACEAE A. Richard 1822 (Touch-me-not Family) A family of 2 genera and 850-1000 species, primarily of the Old World tropics. References: Fischer in Kubitzki (2004). Impatiens Linnaeus (Jewelweed, Touch-me-not, Snapweed, Balsam) A genus of 850-1000 species, herbs and subshrubs, primarily tropical and north temperate Old World. References: Fischer in Kubitzki (2004). 1 Corolla purple, pink, or white; plants 3-6 (-8) dm tall; stems puberulent or glabrous; [cultivated alien, rarely escaped]. 2 Sepal spur strongly recurved; stems puberulent..............................................................................................I. balsamina 2 Sepal spur slightly -

The Boechera Genus As a Resource for Apomixis Research
Zurich Open Repository and Archive University of Zurich Main Library Strickhofstrasse 39 CH-8057 Zurich www.zora.uzh.ch Year: 2019 The Boechera Genus as a Resource for Apomixis Research Brukhin, Vladimir ; Osadtchiy, Jaroslaw V ; Florez-Rueda, Ana Marcela ; Smetanin, Dmitry ; Bakin, Evgeny ; Nobre, Margarida Sofia ; Grossniklaus, Ueli Abstract: he genera Boechera (A. Löve et D. Löve) and Arabidopsis, the latter containing the model plant Arabidopsis thaliana, belong to the same clade within the Brassicaceae family. Boechera is the only among the more than 370 genera in the Brassicaceae where apomixis is well documented. Apomixis refers to the asexual reproduction through seed, and a better understanding of the underlying mechanisms has great potential for applications in agriculture. The Boechera genus currently includes 110 species (of which 38 are reported to be triploid and thus apomictic), which are distributed mostly in the North America. The apomictic lineages of Boechera occur at both the diploid and triploid level and show signs of a hybridogenic origin, resulting in a modification of their chromosome structure, as reflected by alloploidy, aneuploidy, substitutions of homeologous chromosomes, and the presence of aberrant chromosomes. In this review, we discuss the advantages of the Boechera genus to study apomixis, consider its modes of reproduction as well as the inheritance and possible mechanisms controlling apomixis. We also consider population genetic aspects and a possible role of hybridization at the origin of apomixis in Boechera. The molecular tools available to study Boechera, such as transformation techniques, laser capture microdissection, analysis of transcriptomes etc. are also discussed. We survey available genome assemblies of Boechera spp. -

Super-Resolution Ribosome Profiling Reveals Unannotated Translation
Super-resolution ribosome profiling reveals PNAS PLUS unannotated translation events in Arabidopsis Polly Yingshan Hsua, Lorenzo Calviellob,c, Hsin-Yen Larry Wud,1, Fay-Wei Lia,e,f,1, Carl J. Rothfelse,f, Uwe Ohlerb,c, and Philip N. Benfeya,g,2 aDepartment of Biology, Duke University, Durham, NC 27708; bBerlin Institute for Medical Systems Biology, Max Delbrück Center for Molecular Medicine, 13125 Berlin, Germany; cDepartment of Biology, Humboldt Universität zu Berlin, 10099 Berlin, Germany; dBioinformatics Research Center and Department of Statistics, North Carolina State University, Raleigh, NC 27695; eUniversity Herbarium, University of California, Berkeley, CA 94720; fDepartment of Integrative Biology, University of California, Berkeley, CA 94720; and gHoward Hughes Medical Institute, Duke University, Durham, NC 27708 Contributed by Philip N. Benfey, September 13, 2016 (sent for review June 30, 2016; reviewed by Pam J. Green and Albrecht G. von Arnim) Deep sequencing of ribosome footprints (ribosome profiling) maps and contaminants. Several metrics associated with translation have and quantifies mRNA translation. Because ribosomes decode mRNA been exploited (11), for example, the following: (i) ribosomes re- every 3 nt, the periodic property of ribosome footprints could be lease after encountering a stop codon (9), (ii) local enrichment of used to identify novel translated ORFs. However, due to the limited footprints within the predicted ORF (4, 13), (iii) ribosome footprint resolution of existing methods, the 3-nt periodicity is observed length distribution (7), and (iv) 3-nt periodicity displayed by trans- mostly in a global analysis, but not in individual transcripts. Here, we lating ribosomes (2, 6, 10, 14, 15). Among these features, some work report a protocol applied to Arabidopsis that maps over 90% of the well in distinguishing groups of coding vs. -

A Simple Metabolic Switch May Activate Apomixis in <I>Arabidopsis
Utah State University DigitalCommons@USU All Graduate Theses and Dissertations Graduate Studies 12-2018 A Simple Metabolic Switch May Activate Apomixis in Arabidopsis thaliana David Alan Sherwood Utah State University Follow this and additional works at: https://digitalcommons.usu.edu/etd Part of the Agronomy and Crop Sciences Commons, and the Molecular Biology Commons Recommended Citation Sherwood, David Alan, "A Simple Metabolic Switch May Activate Apomixis in Arabidopsis thaliana" (2018). All Graduate Theses and Dissertations. 7409. https://digitalcommons.usu.edu/etd/7409 This Dissertation is brought to you for free and open access by the Graduate Studies at DigitalCommons@USU. It has been accepted for inclusion in All Graduate Theses and Dissertations by an authorized administrator of DigitalCommons@USU. For more information, please contact [email protected]. A SIMPLE METABOLIC SWITCH MAY ACTIVATE APOMIXIS IN ARABIDOPSIS THALIANA by David Alan Sherwood A dissertation submitted in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY in Plant Science – Molecular Biology Approved: _________________________ _________________________ John G. Carman Ph.D. Bruce Bugbee Ph.D. Major Professor Committee Member _________________________ _________________________ Lee Rickords Ph.D. Steve Larson Ph.D. Committee Member Committee Member _________________________ __________________________ Zhongde Wang Ph.D. Laurens H. Smith Ph.D. Committee Member Interim Vice President for Research and Dean of the School of Graduate Studies UTAH STATE UNIVERSITY Logan, Utah 2018 ii COPYRIGHT © David Sherwood 2018 All Rights Reserved iii ABSTRACT A Simple Metabolic Switch May Activate Apomixis in Arabidopsis thaliana by David Alan Sherwood, Doctor of Philosophy Utah State University, 2018 Major Professor: Dr. John G. -

Mandakova TPC2010, Suppl Mat.Pdf
Supplemental Data. Mandáková et al. (2010). Plant Cell 10.1105/tpc.110.074526 Supplemental Figure 1. A Three-Way Comparison of the Relative Position of Corresponding Synteny Blocks of Stenopetalum nutans (SN), S. lineare (SL) and Ballantinia antipoda (BA) Relative to the Reference Ancestral Crucifer Karyotype (ACK). In the main panel of the image, each of the three modern karyotypes is presented in a radial layout. Within each of the three karyotypes, ideograms are ordered and oriented in the outward direction (corresponding to the same counter-clockwise scale progression of Figure 3). Each line connects a pair of genomic positions on two different modern genomes that are syntenically related to the same genomic block in the ACK. For example, the light-blue line at the top of the figure between SL and SN corresponds to synteny with the genomic block U2 on AK7. Each of the eight small panels shows synteny relationships between the three modern genomes for a specific ancestral chromosome (AK1-8). Supplemental Data. Mandáková et al. (2010). Plant Cell 10.1105/tpc.110.074526 Supplemental Figure 2. The Unique Rearrangement of the AK8(#1)-like Homoeologue Shared by All Analyzed Species. This rearrangement was mediated by two subsequent paracentric inversions involving two thirds of genomic block W1 and a major part of X1. In S. lineare, block V1 underwent a secondary translocation to another chromosome. The rearrangement not shown for Arabidella eremigena and Blennodia canescens. Supplemental Data. Mandáková et al. (2010). Plant Cell 10.1105/tpc.110.074526 Supplemental Figure 3. Phylogeny of the Malate Synthase (MS) (TrN + Γ + I) Showing the Position of Sequences from the Australian Species (in Bold) in the Context of Other Brassicaceae Taxa. -

Evolution and Diversification of the Plant Gibberellin Receptor GID1
Evolution and diversification of the plant gibberellin receptor GID1 Hideki Yoshidaa,b, Eiichi Tanimotoc, Takaaki Hiraia, Yohei Miyanoirid,e, Rie Mitania, Mayuko Kawamuraa, Mitsuhiro Takedad,f, Sayaka Takeharaa, Ko Hiranoa, Masatsune Kainoshod,g, Takashi Akagih, Makoto Matsuokaa,1, and Miyako Ueguchi-Tanakaa,1 aBioscience and Biotechnology Center, Nagoya University, Nagoya, 464-8601 Aichi, Japan; bKihara Institute for Biological Research, Yokohama City University, Yokohama, 244-0813 Kanagawa, Japan; cGraduate School of Natural Sciences, Nagoya City University, Nagoya, 467-8501 Aichi, Japan; dStructural Biology Research Center, Graduate School of Science, Nagoya University, Nagoya, 464-8601 Aichi, Japan; eResearch Center for State-of-the-Art Functional Protein Analysis, Institute for Protein Research, Osaka University, Suita, 565-0871 Osaka, Japan; fDepartment of Structural BioImaging, Faculty of Life Sciences, Kumamoto University, 862-0973 Kumamoto, Japan; gGraduate School of Science and Engineering, Tokyo Metropolitan University, Hachioji, 192-0397 Tokyo, Japan; and hGraduate School of Agriculture, Kyoto University, 606-8502 Kyoto, Japan Edited by Mark Estelle, University of California, San Diego, La Jolla, CA, and approved July 10, 2018 (received for review April 9, 2018) The plant gibberellin (GA) receptor GID1 shows sequence similarity erwort Marchantia polymorpha (5–7). Furthermore, Hirano et al. to carboxylesterase (CXE). Here, we report the molecular evolution (5) reported that GID1s in the lycophyte Selaginella moellen- of GID1 from establishment to functionally diverse forms in dorffii (SmGID1s) have unique properties in comparison with eudicots. By introducing 18 mutagenized rice GID1s into a rice angiosperm GID1s: namely, lower affinity to bioactive GAs and gid1 null mutant, we identified the amino acids crucial for higher affinity to inactive GAs (lower specificity). -

Tharp's Blue-Star (2013-2014)
Brack’s Hardwall Cactus Distribution, Habitat, and Status Survey 2015 Natural Heritage New Mexico Report 393 – May, 2016 Brack’s Hardwall Cactus Distribution, Habitat, and Status Survey 20151 Esteban Muldavin,2 Robert Sivinski,3 Mitchell East, Yvonne Chauvin, and Mark Horner Natural Heritage New Mexico, Museum of Southwestern Biology and Department of Biology University of New Mexico, Albuquerque, New Mexico 87131 May 2016 ________________________________________________________________________ Summary Brack’s hardwall cactus (Sclerocactus cloverae subsp. brackii) is a rare plant that occurs on the Nacimiento Formation of northwestern New Mexico. It is listed as a BLM sensitive species and is included on the State of New Mexico list of endangered plant species. Most of the known populations of Brack’s are on lands under Bureau of Land Management (BLM) jurisdiction (about 80%), but it is also present on the Navajo Nation, State of New Mexico Trust Lands, and private property. Portions of the Nacimiento Formation have actively developing oil and gas fields that include the construction of well pads, roads and pipelines that have the potential to significantly negatively impact this species. Yet, the full extent of the species regionally, its habitat requirements, and abundance have not been systematically addressed to support the management of the species. Accordingly, Natural Heritage New Mexico (NHNM) conducted field study in 2015 with two primary objectives: 1) provide a regional assessment of the overall distribution of Sclerocactus cloverae ssp. brackii on the Nacimiento Formation in the context of other closely related Sclerocactus species; and 2 ) describe the potential habitats and relative degree of occupancy by the species within those habitats focusing on the Lybrook area, a major population center for the species and an area of ongoing intensive oil and gas exploration.