Evolution and Diversification of the Plant Gibberellin Receptor GID1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Selfing Syndrome Overshadows Other Differences When Comparing
bioRxiv preprint doi: https://doi.org/10.1101/2020.11.26.398016; this version posted November 27, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 The selfing syndrome overshadows other differences when 2 comparing fitness across Capsella species 3 4 5 Marion Orsucci1, Theofilos Vanikiotis2, Maria Guerrina1, Tianlin Duan1, Sylvain Glémin3, Martin 6 Lascoux1 7 8 1 Department of Ecology and Genetics, Evolutionary Biology Centre and Science for Life 9 Laboratory, Uppsala University, 75236 Uppsala, Sweden 10 2 Department of Biological Applications & Technology, University of Ioannina, Leof. S. 11 Niarchou GR-451 10, Ioannina, Greece 12 3 UMR CNRS 6553 ECOBIO, Campus Beaulieu, bât 14a, p.118, CS 74205, 35042 Rennes, 13 France 14 15 16 Corresponding authors: Martin Lascoux ([email protected]), Marion Orsucci 17 ([email protected]) 18 19 20 Running title: Influence of mating system on life history traits in Capsella spp. 21 22 23 Key words: mating system, ploidy, life history traits, environmental disturbance 24 25 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.11.26.398016; this version posted November 27, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 26 SUMMARY 27 Self-fertilization has recurrently evolved from outcrossing. Self-fertilization provides an advantage 28 in the short-term as individuals do not require a mate to reproduce, but self-fertilization is also 29 associated with both decreased genetic diversity and accumulation of weakly deleterious mutations, 30 which could, however, be alleviated in polyploid selfers. -

Pests, Diseases, and Aridity Have Shaped the Genome of Corymbia Citriodora
Lawrence Berkeley National Laboratory Recent Work Title Pests, diseases, and aridity have shaped the genome of Corymbia citriodora. Permalink https://escholarship.org/uc/item/5t51515k Journal Communications biology, 4(1) ISSN 2399-3642 Authors Healey, Adam L Shepherd, Mervyn King, Graham J et al. Publication Date 2021-05-10 DOI 10.1038/s42003-021-02009-0 Peer reviewed eScholarship.org Powered by the California Digital Library University of California ARTICLE https://doi.org/10.1038/s42003-021-02009-0 OPEN Pests, diseases, and aridity have shaped the genome of Corymbia citriodora ✉ Adam L. Healey 1,2 , Mervyn Shepherd 3, Graham J. King 3, Jakob B. Butler 4, Jules S. Freeman 4,5,6, David J. Lee 7, Brad M. Potts4,5, Orzenil B. Silva-Junior8, Abdul Baten 3,9, Jerry Jenkins 1, Shengqiang Shu 10, John T. Lovell 1, Avinash Sreedasyam1, Jane Grimwood 1, Agnelo Furtado2, Dario Grattapaglia8,11, Kerrie W. Barry10, Hope Hundley10, Blake A. Simmons 2,12, Jeremy Schmutz 1,10, René E. Vaillancourt4,5 & Robert J. Henry 2 Corymbia citriodora is a member of the predominantly Southern Hemisphere Myrtaceae family, which includes the eucalypts (Eucalyptus, Corymbia and Angophora; ~800 species). 1234567890():,; Corymbia is grown for timber, pulp and paper, and essential oils in Australia, South Africa, Asia, and Brazil, maintaining a high-growth rate under marginal conditions due to drought, poor-quality soil, and biotic stresses. To dissect the genetic basis of these desirable traits, we sequenced and assembled the 408 Mb genome of Corymbia citriodora, anchored into eleven chromosomes. Comparative analysis with Eucalyptus grandis reveals high synteny, although the two diverged approximately 60 million years ago and have different genome sizes (408 vs 641 Mb), with few large intra-chromosomal rearrangements. -

Taxa Named in Honor of Ihsan A. Al-Shehbaz
TAXA NAMED IN HONOR OF IHSAN A. AL-SHEHBAZ 1. Tribe Shehbazieae D. A. German, Turczaninowia 17(4): 22. 2014. 2. Shehbazia D. A. German, Turczaninowia 17(4): 20. 2014. 3. Shehbazia tibetica (Maxim.) D. A. German, Turczaninowia 17(4): 20. 2014. 4. Astragalus shehbazii Zarre & Podlech, Feddes Repert. 116: 70. 2005. 5. Bornmuellerantha alshehbaziana Dönmez & Mutlu, Novon 20: 265. 2010. 6. Centaurea shahbazii Ranjbar & Negaresh, Edinb. J. Bot. 71: 1. 2014. 7. Draba alshehbazii Klimeš & D. A. German, Bot. J. Linn. Soc. 158: 750. 2008. 8. Ferula shehbaziana S. A. Ahmad, Harvard Pap. Bot. 18: 99. 2013. 9. Matthiola shehbazii Ranjbar & Karami, Nordic J. Bot. doi: 10.1111/j.1756-1051.2013.00326.x, 10. Plocama alshehbazii F. O. Khass., D. Khamr., U. Khuzh. & Achilova, Stapfia 101: 25. 2014. 11. Alshehbazia Salariato & Zuloaga, Kew Bulletin …….. 2015 12. Alshehbzia hauthalii (Gilg & Muschl.) Salariato & Zuloaga 13. Ihsanalshehbazia Tahir Ali & Thines, Taxon 65: 93. 2016. 14. Ihsanalshehbazia granatensis (Boiss. & Reuter) Tahir Ali & Thines, Taxon 65. 93. 2016. 15. Aubrieta alshehbazii Dönmez, Uǧurlu & M.A.Koch, Phytotaxa 299. 104. 2017. 16. Silene shehbazii S.A.Ahmad, Novon 25: 131. 2017. PUBLICATIONS OF IHSAN A. AL-SHEHBAZ 1973 1. Al-Shehbaz, I. A. 1973. The biosystematics of the genus Thelypodium (Cruciferae). Contrib. Gray Herb. 204: 3-148. 1977 2. Al-Shehbaz, I. A. 1977. Protogyny, Cruciferae. Syst. Bot. 2: 327-333. 3. A. R. Al-Mayah & I. A. Al-Shehbaz. 1977. Chromosome numbers for some Leguminosae from Iraq. Bot. Notiser 130: 437-440. 1978 4. Al-Shehbaz, I. A. 1978. Chromosome number reports, certain Cruciferae from Iraq. -
Eutrema Salsugineum (Cruciferae) New to Mexico: a Surprising Generic
A peer-reviewed open-access journal PhytoKeys 76:Eutrema 13–21 (2017) salsugineum (Cruciferae) new to Mexico: a surprising generic record... 13 doi: 10.3897/phytokeys.76.9731 SHORT COMMUNICATION http://phytokeys.pensoft.net Launched to accelerate biodiversity research Eutrema salsugineum (Cruciferae) new to Mexico: a surprising generic record for the flora of Middle America Dmitry A. German1,2, Marcus A. Koch1 1 Department of Biodiversity and Plant Systematics, Centre for Organismal Studies (COS) Heidelberg, Hei- delberg University, Im Neuenheimer Feld 345, D-69120 Heidelberg, Germany 2 South-Siberian Botanical Garden, Altai State University, Lenin Str. 61, 656049 Barnaul, Russia Corresponding author: Dmitry A. German ([email protected]) Academic editor: P. de Lange | Received 9 November 2016 | Accepted 12 December 2016 | Published 5 January 2017 Citation: German DA, Koch MA (2017) Eutrema salsugineum (Cruciferae) new to Mexico: a surprising generic record for the flora of Middle America. PhytoKeys 76: 13–21. https://doi.org/10.3897/phytokeys.76.9731 Abstract The paper reports Eutrema salsugineum as a novelty to the flora of Mexico and Middle America in general. The finding stands ca. 1600 km apart from the closest known locality in the Rocky Mountains of Colora- do, USA. The species is considered native to NW Mexico and its late discovery in the region is presumably explained by its tiny habit, early flowering time, and subephemeral life cycle. The phylogenetic position of this Mexican population in a haplotype network based on the chloroplast DNA fragment psbA-trnH confirms this hypothesis and also suggests, in contrast to the previously held viewpoint, multiple coloniza- tions of North American continent from Asia. -

Is the Endangered Species Act in Danger?
Aqui egza• Newsletter of the Colorado Native Plant Society " ... dedicated to the appreciation and conservation of the Colorado native flora" ......... ..... .. ~ ........... ," .. ,' .... ", ., ..... ', .. " .. -.". ","' ....... " ... _, qtl.ty> •• t.h.·rQI.J.g.h •• ·} ••• [).~.C~nlt>e," 1.·9.93··· Is The Endangered Species Act In Danger? Nina Williams under the Act are those fortunate enough to Furthermore, unlike wildlife and fish, only Legislative Affairs Committee reside on federal land, those endangered by plant species or sub-species, not populations, a project receiving federal funds, or those are eligible for protection under the Act. The battle over our nation's strongest plant species afforded protection by state This means that unless a plant is imperiled environmental law , the Endangered Species law. This last provision offers no help in throughout its entire range, it can not be Act of 1973 (ESA, the Act), is heating up in Colorado where the only plant protected by protected under the Act. In contrast, a Congress. The ESA is now 20 years old and state law is the ubiquitous state flower, the threatened or endangered population of ~ up for re-authorization in the next session of Colorado columbine (Aqui/egia caerulea). animals can be protected under the Act even the United States Congress. It is no surprise Colorado state law provides absolutely NO if viable populations of the same species that the Act is facing strong opposition from protection for any other plant species on exist elsewhere. conservative private property rights private land, including the 88 species advocates such as the Wise Use Movement. identified by the Colorado Natural Heritage Inequitable treatment ofplants has been the High profile struggles such as old growth Program as rare. -

IEG News June Edition
June 2019 IEG News June edition News from the Head of the Department I wish everyone at the department a nice summer - relaxing vacations, and successful work in the field or elsewhere. Thank you all for contributions during the past academic year! For the upcoming fall, don’t forget the following important events: - The dean of biology has invited all senior scientists (“PIs”) to strategic discussions about biology at Uppsala University on August 28 - see mail sent by Staffan Svärd on June 5 for details and reg- istration. - The annual biology teacher days, August 22-23 - see mail from Henning Blom on June 3 for de- tails and registration. - The annual IEG Day will take place 8th of November - a full day of information, interactions, and discussion on topics of importance to the success of IEG as an excellent academic environment. There will also be research presentations, and a party. Old colleague moves on to new challenges Stefan Bertilsson leaves us for a professor position at the Department of Aquatic Sciences and As- sessment at SLU. Good luck to Stefan in his new job, and we look forward to coming inter-univer- sity cooperation! At the farewell celebration, we equipped him with some relevant tools for his fu- ture at the agricultural university. Stefan will still be around to some extent to finalize supervision. Photos courtesy of Lars Tranvik News from the administration Travelling on a business trip? Business trips must be booked through Lingmerth’s travel agency, which is the university’s pro- cured supplier, read more on the Employee Portal (Medarbetarportalen). -

Wild Crop Relatives: Genomic and Breeding Resources: Forest Trees
Wild Crop Relatives: Genomic and Breeding Resources . Chittaranjan Kole Editor Wild Crop Relatives: Genomic and Breeding Resources Forest Trees Editor Prof. Chittaranjan Kole Director of Research Institute of Nutraceutical Research Clemson University 109 Jordan Hall Clemson, SC 29634 [email protected] ISBN 978-3-642-21249-9 e-ISBN 978-3-642-21250-5 DOI 10.1007/978-3-642-21250-5 Springer Heidelberg Dordrecht London New York Library of Congress Control Number: 2011922649 # Springer-Verlag Berlin Heidelberg 2011 This work is subject to copyright. All rights are reserved, whether the whole or part of the material is concerned, specifically the rights of translation, reprinting, reuse of illustrations, recitation, broadcasting, reproduction on microfilm or in any other way, and storage in data banks. Duplication of this publication or parts thereof is permitted only under the provisions of the German Copyright Law of September 9, 1965, in its current version, and permission for use must always be obtained from Springer. Violations are liable to prosecution under the German Copyright Law. The use of general descriptive names, registered names, trademarks, etc. in this publication does not imply, even in the absence of a specific statement, that such names are exempt from the relevant protective laws and regulations and therefore free for general use. Cover design: deblik, Berlin Printed on acid-free paper Springer is part of Springer Science+Business Media (www.springer.com) Dedication Dr. Norman Ernest Borlaug,1 the Father of Green Revolution, is well respected for his contribu- tions to science and society. There was or is not and never will be a single person on this Earth whose single-handed service to science could save millions of people from death due to starvation over a period of over four decades like Dr. -
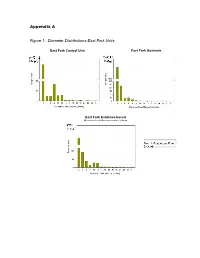
Appendix a Figure 1: Diameter Distributions-East Fork Units
Appendix A Figure 1: Diameter Distributions-East Fork Units East Fork Control Unit East Fork Goshawk Guidelines Unit East Fork Evidence-based Ecological Restoration Unit Figure 2: Diameter Distributions-Redondo Units Redondo Control Unit Redondo Goshawk Guidelines Unit Redondo Evidence-based Ecological Restoration Unit Figure 3: FlamMap Simulated Crown Fire Behavior: East Fork Project Area 20 mph Southwest Wind 25 mph 30 mph 35 mph 40 mph Map Legend Crown Fire Activity N No crown fire Passive crown activity fire (torching) Active crown fire (spreading) Important Note: Expected Crown Fire Behavior by Percent The 97th percentile of low fuel of Project Area moistures for June from 34 No Crown Passive years of data from the Kaibab Fire Crown Fire Active National Forest, Arizona were used in all simulations. This Windspeed Activity (torching) Crown Fire 20 79.9 0.9 19.2 input represents potential 25 71.2 1.9 26.9 extremes of fire weather during the peak of the fire 30 64.9 2.9 32.3 season, not average fire 35 57.3 4.7 38.0 season conditions. 40 48.7 9.4 42.0 Figure 4: FlamMap Simulated Crown Fire Behavior: Redondo Project Area 20 mph Southwest Wind 25 mph 30 mph 35 mph 40 mph Map Legend Crown Fire Activity N Highway 4 Valles Caldera No crown fire Passive crown activity entrance road fire (torching) Active crown fire (spreading) Important Note: Expected Crown Fire Behavior by Percent The 97th percentile of low fuel of Project Area moistures for June from 34 Passive years of data from the Kaibab No Crown Crown Fire Active Crown National Forest, Arizona were Windspeed Fire Activity (torching) Fire used in all simulations. -

Wasabi – Eutrema Japonicum
Did You Know? Wasabi – Eutrema japonicum • Is in the Brassicaceae family, the same family as horseradish, mustard, cabbage and kale • Japanese horseradish is a common name (although it is not in the same genus as horseradish) • Prepared wasabi comes from grating the rhizome of the Eutrema japonicum plant • Most commonly sold as a dried powder or as a paste in a tube • Originated in Japan, and can be traced back to 794 CE • The condiment sold as wasabi often contains no wasabi at all, but is rather a mixture of horseradish, mustard and food coloring • Leaves, rhizomes, stems (petioles) and flowers of the wasabi plant are edible • Grate the wasabi rhizome just before serving as it loses flavor 10- 15 minutes after it has been prepared • Traditionally, the grater used to prepare wasabi was made from a piece of dry sharkskin stapled to a wooden paddle. Grating was done by pressing the rhizome onto the paddle and moved in a circular motion until a paste formed. • Studied for medicinal uses, including as an anti-inflammatory, anti-bacterial and to reduce risk of osteoporosis, heart disease, and cancer • Is high in antioxidants, vitamin C and contains some vitamin A and iron • Wasabi grows in cool, shady conditions in temperate regions (45-70˚F), preferring high humidity during the summer months and ample water. • Wasabi requires 18 months and up to three years of growth to reach maturity • Commercially, wasabi is grown in northern Japan, parts of China, Taiwan, Korea and New Zealand. The right climate balance for successful wasabi growth can be found in North America in the rain forests on the Oregon Coast and parts of the Blue Ridge Mountains in North Carolina and Tennessee. -

Genome 2Fractional WGD 2412.6 Γ 1595.2 ): 0.17 0.51 Ε ): 0.64 • 233.6 136.8 0.02
Where does biological order come from? Gavin Conant Bioinformatics Research Center Biological Sciences Program in Genetics [email protected] conantlab.org Tracking flipprob.:0.0081 Fixation rate( preservationrate( Genome 2fractional WGD 2412.6 γ 1595.2 ): 0.17 0.51 ε ): 0.64 • 233.6 136.8 0.02 0.09 133.0 84.8 0.08 105.7 0.06 31.2 187.4 35.0 Metagenomics 312.3 281.2 188.7 298.0 410.1 192.6 0.16 195.8 0.17 997.0 282.0 0.17 388.0 608.2 0.54 0.24 258.8 0.23 Thellungiella halophila Thellungiella halophila Arabidopsis thaliana Arabidopsis thaliana Aethionema arabicum Eutrema parvulum Eutrema parvulum Aethionema arabicum Arabidopsis lyrata Arabidopsis lyrata Capsella rubella Capsella rubella 3189.3 3095.4 3126.4 2203.4 2033.5 2096.5 2098.7 2036.6 2040.6 3409.6 3131.5 3221.5 AA32G00445 Tp4g15760 20180168 10023932 AT2G33430 DAL 0.999 Tp1g03280 20187201 10012323 AT1G04450 AA32G00441 Tp4g15800 20179724 10025564 4001438 RIC1 0.998 AA19G00112 Tp1g03240 20188475 10009108 100399 AT1G04420 0.999 from PG#1 All singlecopy AA19G00113 Tp1g03230 20188581 AT1G04410 C-NAD-MDH1, AA32G00436 Tp2g08140 20208201 10026756 AT5G43330 C-NAD-MDH2 0.997 Tp1g03220 20187279 10008610 AT1G04400 AT-PHH1 AA32G00435 Tp2g08150 20208192 0.997 Tp1g03180 20187678 10011057 100393 ERF14 AA32G00429 Tp2g08290 20207965 10028227 800707 ERF96 0.995 AA19G00116 Tp1g03170 20188613 10012007 100392 AA32G00428 Tp2g08300 20207948 10028453 AT5G43420 AT5G43420 0.994 from PG#2 All singlecopy AA19G00118 Tp1g03150 20188264 10010342 100390 AT1G04340 AA32G00426 Tp2g08340 20208037 10027196 AT5G43460 0.994 -

Capsella Bursa-Pastoris ) – Establishment of a New Model System
Characterisation of the natural homeotic variety Stamenoid petals (Spe ) in the Shepherd´s Purse ( Capsella bursa-pastoris ) – Establishment of a new model system Dissertation zur Erlangung des akademischen Grades doctor rerum naturalium (Dr. rer. nat.) vorgelegt dem Rat der Biologisch-Pharmazeutischen Fakultät der Friedrich-Schiller-Universität Jena von Diplom-Biologin Pia Nutt Geboren am 9. Juni 1973 in Paderborn Gutachter 1. Prof. Dr. Günter Theißen (Jena) 2. Prof. Dr. Ralf Oelmüller (Jena) 3. PD Dr. Stefan Gleissberg (Ohio, USA) Tag der öffentlichen Verteidigung: Donnerstag, den 18. Dezember 2008 Meinen Eltern und Jorge Table of contents Table of contents 1 Introduction …………………………………………………………………….. 3 1.1 About homeosis………………………………………………………….. 3 1.2 Developmental genetics of floral homeotic mutants ……………………. 5 1.3 The role of homeotic mutants in the evolution of flowers……………….. 7 1.4 A floral homeotic variant of C. bursa-pastoris helps investigating the evolutionary role of homeosis in plants………………………………….. 8 1.5 Capsella bursa-pastoris as a model species……………………………… 10 1.6 Aims of this work………………………………………………………… 11 2 Overview of the manuscripts …………………………………………………… 14 3 Manuscript I ……………………………………………………………………... 16 P. Nutt , J. Ziermann, M. Hintz, B. Neuffer, and G. Theißen (2006): Capsella as a model system to study the evolutionary relevance of floral homeotic mutants. Plant Systematics and Evolution 259, pp 217-235. 4 Manuscript II ……………………………………………………………………. 36 P. Nutt 1, Janine Ziermann 1 and G. Theißen (submitted to The Plant Cell on May 7, 2008) Ectopic expression and co-segregation of an AGAMOUS orthologue in Stamenoid petals , a natural homeotic floral variant of Capsella bursa-pastoris. ( 1 These authors contributed equally to this work) 5 Manuscript III …………………………………………………………………… 84 C. -

Washington Flora Checklist a Checklist of the Vascular Plants of Washington State Hosted by the University of Washington Herbarium
Washington Flora Checklist A checklist of the Vascular Plants of Washington State Hosted by the University of Washington Herbarium The Washington Flora Checklist aims to be a complete list of the native and naturalized vascular plants of Washington State, with current classifications, nomenclature and synonymy. The checklist currently contains 3,929 terminal taxa (species, subspecies, and varieties). Taxa included in the checklist: * Native taxa whether extant, extirpated, or extinct. * Exotic taxa that are naturalized, escaped from cultivation, or persisting wild. * Waifs (e.g., ballast plants, escaped crop plants) and other scarcely collected exotics. * Interspecific hybrids that are frequent or self-maintaining. * Some unnamed taxa in the process of being described. Family classifications follow APG IV for angiosperms, PPG I (J. Syst. Evol. 54:563?603. 2016.) for pteridophytes, and Christenhusz et al. (Phytotaxa 19:55?70. 2011.) for gymnosperms, with a few exceptions. Nomenclature and synonymy at the rank of genus and below follows the 2nd Edition of the Flora of the Pacific Northwest except where superceded by new information. Accepted names are indicated with blue font; synonyms with black font. Native species and infraspecies are marked with boldface font. Please note: This is a working checklist, continuously updated. Use it at your discretion. Created from the Washington Flora Checklist Database on September 17th, 2018 at 9:47pm PST. Available online at http://biology.burke.washington.edu/waflora/checklist.php Comments and questions should be addressed to the checklist administrators: David Giblin ([email protected]) Peter Zika ([email protected]) Suggested citation: Weinmann, F., P.F. Zika, D.E. Giblin, B.