Systematic Identification of Human Snps Affecting Regulatory Element
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Screening and Identification of Key Biomarkers in Clear Cell Renal Cell Carcinoma Based on Bioinformatics Analysis
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.21.423889; this version posted December 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Screening and identification of key biomarkers in clear cell renal cell carcinoma based on bioinformatics analysis Basavaraj Vastrad1, Chanabasayya Vastrad*2 , Iranna Kotturshetti 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karanataka, India. 3. Department of Ayurveda, Rajiv Gandhi Education Society`s Ayurvedic Medical College, Ron, Karnataka 562209, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India bioRxiv preprint doi: https://doi.org/10.1101/2020.12.21.423889; this version posted December 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Clear cell renal cell carcinoma (ccRCC) is one of the most common types of malignancy of the urinary system. The pathogenesis and effective diagnosis of ccRCC have become popular topics for research in the previous decade. In the current study, an integrated bioinformatics analysis was performed to identify core genes associated in ccRCC. An expression dataset (GSE105261) was downloaded from the Gene Expression Omnibus database, and included 26 ccRCC and 9 normal kideny samples. Assessment of the microarray dataset led to the recognition of differentially expressed genes (DEGs), which was subsequently used for pathway and gene ontology (GO) enrichment analysis. -

Molecular Effects of Isoflavone Supplementation Human Intervention Studies and Quantitative Models for Risk Assessment
Molecular effects of isoflavone supplementation Human intervention studies and quantitative models for risk assessment Vera van der Velpen Thesis committee Promotors Prof. Dr Pieter van ‘t Veer Professor of Nutritional Epidemiology Wageningen University Prof. Dr Evert G. Schouten Emeritus Professor of Epidemiology and Prevention Wageningen University Co-promotors Dr Anouk Geelen Assistant professor, Division of Human Nutrition Wageningen University Dr Lydia A. Afman Assistant professor, Division of Human Nutrition Wageningen University Other members Prof. Dr Jaap Keijer, Wageningen University Dr Hubert P.J.M. Noteborn, Netherlands Food en Consumer Product Safety Authority Prof. Dr Yvonne T. van der Schouw, UMC Utrecht Dr Wendy L. Hall, King’s College London This research was conducted under the auspices of the Graduate School VLAG (Advanced studies in Food Technology, Agrobiotechnology, Nutrition and Health Sciences). Molecular effects of isoflavone supplementation Human intervention studies and quantitative models for risk assessment Vera van der Velpen Thesis submitted in fulfilment of the requirements for the degree of doctor at Wageningen University by the authority of the Rector Magnificus Prof. Dr M.J. Kropff, in the presence of the Thesis Committee appointed by the Academic Board to be defended in public on Friday 20 June 2014 at 13.30 p.m. in the Aula. Vera van der Velpen Molecular effects of isoflavone supplementation: Human intervention studies and quantitative models for risk assessment 154 pages PhD thesis, Wageningen University, Wageningen, NL (2014) With references, with summaries in Dutch and English ISBN: 978-94-6173-952-0 ABSTRact Background: Risk assessment can potentially be improved by closely linked experiments in the disciplines of epidemiology and toxicology. -

Identification of Bleomycin and Radiation-Induced Pulmonary Fibrosis Susceptibility Genes in Mice Anne-Marie Lemay Department Of
Identification of bleomycin and radiation-induced pulmonary fibrosis susceptibility genes in mice Anne-Marie Lemay Department of Human Genetics McGill University, Montréal February 4th, 2010 A thesis submitted to McGill University in partial fulfilment of the requirements of the degree of Doctor of Philosophy © Anne-Marie Lemay 2010 Comme il est profond, ce mystère de l’Invisible ! Nous ne pouvons le sonder avec nos sens misérables, avec nos yeux qui ne savent apercevoir ni le trop petit, ni le trop grand, ni le trop près, ni le trop loin, ni les habitants d’une étoile, ni les habitants d’une goutte d’eau… Guy de Maupassant Le Horla ii Table of contents Table of contents ................................................................................................... iii Abstract...................................................................................................................vi Résumé ................................................................................................................ viii Acknowledgments................................................................................................... x Abbreviations........................................................................................................ xii Original contributions to knowledge...................................................................xiv Author contribution to research...........................................................................xv List of figures ........................................................................................................ -

Cov-2 Infection in the Small Intestine
Gastroenterology and Hepatology From Bed to Bench. ORIGINAL ARTICLE ©2020 RIGLD, Research Institute for Gastroenterology and Liver Diseases Investigating the human protein-host protein interactome of SARS- CoV-2 infection in the small intestine Mahmoud Khodadoost1, Zahra Niknam2, Masoumeh Farahani2, Mohammadreza Razzaghi3, Mohsen Norouzinia4 1Department of Traditional Medicine, School of Traditional Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran 2Proteomics Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran 3Laser Application in Medical Sciences Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran 4 Gastroenterology and Liver Diseases Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran ABSTRACT Aim: The present study aimed to identify human protein–host protein interactions of SARS-CoV-2 infection in the small intestine to discern the potential mechanisms and gain insights into the associated biomarkers and treatment strategies. Background: Deciphering the tissue and organ interactions of the SARS-CoV-2 infection can be important to discern the potential underlying mechanisms. In the present study, we investigated the human protein–host protein interactions in the small intestine. Methods: Public databases and published works were used to collect data related to small intestine tissue and SARS-CoV-2 infection. We constructed a human protein-protein interaction (PPI) network and showed interactions of host proteins in the small intestine. Associated modules, biological processes, functional pathways, regulatory transcription factors, disease ontology categories, and possible drug candidates for therapeutic targets were identified. Results: Thirteen primary protein neighbors were found for the SARS-CoV-2 receptor ACE2. ACE2 and its four partners were observed in a highly clustered module; moreover, 8 host proteins belonged to this module. -

Aminopeptidase Expression in Multiple Myeloma Associates with Disease Progression and Sensitivity to Melflufen
cancers Article Aminopeptidase Expression in Multiple Myeloma Associates with Disease Progression and Sensitivity to Melflufen Juho J. Miettinen 1,† , Romika Kumari 1,†, Gunnhildur Asta Traustadottir 2 , Maiju-Emilia Huppunen 1, Philipp Sergeev 1, Muntasir M. Majumder 1, Alexander Schepsky 2 , Thorarinn Gudjonsson 2 , Juha Lievonen 3, Despina Bazou 4, Paul Dowling 5, Peter O‘Gorman 4, Ana Slipicevic 6, Pekka Anttila 3, Raija Silvennoinen 3 , Nina N. Nupponen 6, Fredrik Lehmann 6 and Caroline A. Heckman 1,* 1 Institute for Molecular Medicine Finland-FIMM, HiLIFE–Helsinki Institute of Life Science, iCAN Digital Precision Cancer Medicine Flagship, University of Helsinki, 00290 Helsinki, Finland; juho.miettinen@helsinki.fi (J.J.M.); romika.kumari@helsinki.fi (R.K.); maiju-emilia.huppunen@helsinki.fi (M.-E.H.); philipp.sergeev@helsinki.fi (P.S.); muntasir.mamun@helsinki.fi (M.M.M.) 2 Stem Cell Research Unit, Biomedical Center, University of Iceland, 101 Reykjavik, Iceland; [email protected] (G.A.T.); [email protected] (A.S.); [email protected] (T.G.) 3 Department of Hematology, Helsinki University Hospital Comprehensive Cancer Center, 00290 Helsinki, Finland; juha.lievonen@hus.fi (J.L.); pekka.anttila@hus.fi (P.A.); raija.silvennoinen@helsinki.fi (R.S.) 4 Department of Hematology, Mater Misericordiae University Hospital, D07 Dublin, Ireland; [email protected] (D.B.); [email protected] (P.O.) 5 Department of Biology, Maynooth University, National University of Ireland, Citation: Miettinen, J.J.; Kumari, R.; W23 F2H6 Maynooth, Co. Kildare, Ireland; [email protected] Traustadottir, G.A.; Huppunen, M.-E.; 6 Oncopeptides AB, 111 53 Stockholm, Sweden; [email protected] (A.S.); Sergeev, P.; Majumder, M.M.; [email protected] (N.N.N.); [email protected] (F.L.) Schepsky, A.; Gudjonsson, T.; * Correspondence: caroline.heckman@helsinki.fi; Tel.: +358-50-415-6769 † These authors equally contributed to this work. -

Omics Profile Interpretation on Molecular Interaction
Die approbierte Originalversion dieser Dissertation ist an der Hauptbibliothek der Technischen Universität Wien aufgestellt (http://www.ub.tuwien.ac.at). The approved original version of this thesis is available at the main library of the Vienna University of Technology (http://www.ub.tuwien.ac.at/englweb/). Omics profile interpretation on molecular interaction graphs DISSERTATION zur Erlangung des akademischen Grades Doktor der technischen Wissenschaften eingereicht von Raul Fechete Matrikelnummer 0225871 an der Fakultät für Informatik der Technischen Universität Wien Betreuung: Univ.-Prof. Dr. Rudolf Freund Univ.-Doz. Dr. Bernd Mayer Diese Dissertation haben begutachtet: (Univ.-Prof. Dr. Rudolf Freund) (Univ.-Doz. Dr. Bernd Mayer) Wien, 01.10.2012 (Raul Fechete) Technische Universität Wien A-1040 Wien Karlsplatz 13 Tel. +43-1-58801-0 www.tuwien.ac.at Omics profile interpretation on molecular interaction graphs DISSERTATION submitted in partial fulfillment of the requirements for the degree of Doktor der technischen Wissenschaften by Raul Fechete Registration Number 0225871 to the Faculty of Informatics at the Vienna University of Technology Advisors: Univ.-Prof. Dr. Rudolf Freund Univ.-Doz. Dr. Bernd Mayer The dissertation has been reviewed by: (Univ.-Prof. Dr. Rudolf Freund) (Univ.-Doz. Dr. Bernd Mayer) Wien, 01.10.2012 (Raul Fechete) Technische Universität Wien A-1040 Wien Karlsplatz 13 Tel. +43-1-58801-0 www.tuwien.ac.at Erklärung zur Verfassung der Arbeit Raul Fechete Pilgerimgasse 25/13, 1150 Wien Hiermit erkläre ich, dass ich diese Arbeit selbständig verfasst habe, dass ich die verwende- ten Quellen und Hilfsmittel vollständig angegeben habe und dass ich die Stellen der Arbeit - einschließlich Tabellen, Karten und Abbildungen -, die anderen Werken oder dem Internet im Wortlaut oder dem Sinn nach entnommen sind, auf jeden Fall unter Angabe der Quelle als Ent- lehnung kenntlich gemacht habe. -

X‑Linked Congenital Ptosis and Associated Intellectual Disability
Hum Genet (2014) 133:625–638 DOI 10.1007/s00439-013-1403-3 ORIGINAL INVESTIGATION X‑linked congenital ptosis and associated intellectual disability, short stature, microcephaly, cleft palate, digital and genital abnormalities define novel Xq25q26 duplication syndrome R. S. Møller · L. R. Jensen · S. M. Maas · J. Filmus · M. Capurro · C. Hansen · C. L. M. Marcelis · K. Ravn · J. Andrieux · M. Mathieu · M. Kirchhoff · O. K. Rødningen · N. de Leeuw · H. G. Yntema · G. Froyen · J. Vandewalle · K. Ballon · E. Klopocki · S. Joss · J. Tolmie · A. C. Knegt · A. M. Lund · H. Hjalgrim · A. W. Kuss · N. Tommerup · R. Ullmann · A. P. M. de Brouwer · P. Strømme · S. Kjaergaard · Z. Tümer · T. Kleefstra Received: 8 August 2013 / Accepted: 21 November 2013 / Published online: 11 December 2013 © Springer-Verlag Berlin Heidelberg 2013 Abstract Submicroscopic duplications along the long submicroscopic X-chromosome duplications, whereas arm of the X-chromosome with known phenotypic con- duplications in Xq25 and Xq26 have been reported in only sequences are relatively rare events. The clinical features a few cases. Using genome-wide array platforms we iden- resulting from such duplications are various, though they tified overlapping interstitial Xq25q26 duplications rang- often include intellectual disability, microcephaly, short ing from 0.2 to 4.76 Mb in eight unrelated families with stature, hypotonia, hypogonadism and feeding difficulties. in total five affected males and seven affected females.A ll Female carriers are often phenotypically normal or show a affected males shared a common phenotype with intrauter- similar but milder phenotype, as in most cases the X-chro- ine- and postnatal growth retardation and feeding difficul- mosome harbouring the duplication is subject to inactiva- ties in childhood. -
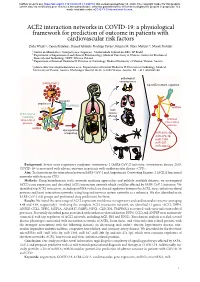
ACE2 Interaction Networks in COVID-19
bioRxiv preprint doi: https://doi.org/10.1101/2020.05.13.094714; this version posted May 14, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. ACE2 interaction networks in COVID-19: a physiological framework for prediction of outcome in patients with cardiovascular risk factors Zofia Wicik1,2, Ceren Eyileten2, Daniel Jakubik2, Rodrigo Pavão1, Jolanta M. Siller-Matula2,3*, Marek Postula2 ¹ Centro de Matemática, Computação e Cognição - Universidade Federal do ABC, SP, Brazil ² Department of Experimental and Clinical Pharmacology, Medical University of Warsaw, Center for Preclinical Research and Technology CEPT, Warsaw, Poland ³ Department of Internal Medicine II, Division of Cardiology, Medical University of Vienna, Vienna, Austria * [email protected]; Department of Internal Medicine II, Division of Cardiology, Medical University of Vienna, Austria. Waehringer Guertel 18-20, A-1090 Vienna, Austria. Tel. +43 1 4040046140 pathological consequences SARS-CoV-2 top ACE2 network regulators activation attachment TMPRSS2 miRNA ACE2 main ACE2 network tissues containing affected virus-related activated proteins virus-related proteins miRNA dysregulation of signaling miR-302c-5p miR-27a-3p miR-1305 miR-587 miR-26b-5p Background: Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection (coronavirus disease 2019; COVID-19) is associated with adverse outcome in patients with cardiovascular disease (CVD). Aim: To characterize the interaction between SARS-CoV-2 and Angiotensin Converting Enzyme 2 (ACE2) functional networks with focus on CVD. -

Breakpoint Determination of X;Autosome Balanced Translocations in Four Patients with Premature Ovarian Failure
Journal of Human Genetics (2011) 56, 156–160 & 2011 The Japan Society of Human Genetics All rights reserved 1434-5161/11 $32.00 www.nature.com/jhg ORIGINAL ARTICLE Breakpoint determination of X;autosome balanced translocations in four patients with premature ovarian failure Akira Nishimura-Tadaki1, Takahito Wada2, Gul Bano3, Karen Gough4, Janet Warner4, Tomoki Kosho5, Noriko Ando6, Haruka Hamanoue1,7, Hideya Sakakibara7, Gen Nishimura8, Yoshinori Tsurusaki1, Hiroshi Doi1, Noriko Miyake1, Keiko Wakui5, Hirotomo Saitsu1, Yoshimitsu Fukushima5, Fumiki Hirahara7 and Naomichi Matsumoto1 Premature ovarian failure (POF) is a disorder characterized by amenorrhea and elevated serum gonadotropins before 40 years of age. As X chromosomal abnormalities are often recognized in POF patients, defects of X-linked gene may contribute to POF. Four cases of POF with t(X;autosome) were genetically analyzed. All the translocation breakpoints were determined at the nucleotide level. Interestingly, COL4A6 at Xq22.3 encoding collagen type IV alpha 6 was disrupted by the translocation in one case, but in the remaining three cases, breakpoints did not involve any X-linked genes. According to the breakpoint sequences, two translocations had microhomology of a few nucleotides and the other two showed insertion of 3–8 nucleotides with unknown origin, suggesting that non-homologous end-joining is related to the formation of all the translocations. Journal of Human Genetics (2011) 56, 156–160; doi:10.1038/jhg.2010.155; published online 9 December 2010 Keywords: COL4A6; critical region; non-homologous end-joining; premature ovarian failure; X;autosome translocation INTRODUCTION translocations. Breakpoint sequences will be presented and discussed Premature ovarian failure (POF) is a disorder characterized by in relation to genes and formation process. -

Genomic and Transcriptomic Characterization of the Human Glioblastoma Cell Line AHOL1
Brazilian Journal of Medical and Biological Research (2021) 54(3): e9571, http://dx.doi.org/10.1590/1414-431X20209571 ISSN 1414-431X Research Article 1/12 Genomic and transcriptomic characterization of the human glioblastoma cell line AHOL1 W.A.S. Ferreira1 00 , C.K.N. Amorim1 00 , R.R. Burbano2,3,4 00 , R.A.R. Villacis5 00 , F.A. Marchi6 00 , T.S. Medina6 00 , M.M.C. de Lima7 00 , and E.H.C. de Oliveira1,8 00 1Laboratório de Cultura de Tecidos e Citogenética, SAMAM, Instituto Evandro Chagas, Ananindeua, PA, Brasil 2Laboratório de Citogenética Humana, Instituto de Ciências Biológicas, Universidade Federal do Pará, Belém, PA, Brasil 3Núcleo de Pesquisas em Oncologia, Hospital Universitário João de Barros Barreto, Belém, PA, Brasil 4Laboratório de Biologia Molecular, Hospital Ophir Loyola, Belém, PA, Brasil 5Departamento de Genética e Morfologia, Instituto de Ciências Biológicas, Universidade de Brasília, Brasília, DF, Brasil 6Centro Internacional de Pesquisa, A.C. Camargo Cancer Center, São Paulo, SP, Brasil 7Instituto de Ciências Biológicas, Faculdade de Biomedicina, Universidade Federal do Pará, Belém, PA, Brasil 8Instituto de Ciências Exatas e Naturais, Faculdade de Ciências Naturais, Universidade Federal do Pará, Belém, PA, Brasil Abstract Cancer cell lines are widely used as in vitro models of tumorigenesis, facilitating fundamental discoveries in cancer biology and translational medicine. Currently, there are few options for glioblastoma (GBM) treatment and limited in vitro models with accurate genomic and transcriptomic characterization. Here, a detailed characterization of a new GBM cell line, namely AHOL1, was conducted in order to fully characterize its molecular composition based on its karyotype, copy number alteration (CNA), and transcriptome profiling, followed by the validation of key elements associated with GBM tumorigenesis. -

Plasma Membrane Protein Identifications from Human Bone Marrow Mesenchymal Stem Cells
Supplementary Material (ESI) for Analytical Methods This journal is (C) The Royal Society of Chemistry 2010 Supplementary Table 1: Plasma Membrane Protein Identifications from Human Bone Marrow Mesenchymal Stem Cells Symbol Entrez Gene Name Type ABCA1 ATP-binding cassette, sub-family A (ABC1), member 1 transporter ABCC1 ATP-binding cassette, sub-family C (CFTR/MRP), member 1 transporter ABCC2 ATP-binding cassette, sub-family C (CFTR/MRP), member 2 transporter ABCC3 ATP-binding cassette, sub-family C (CFTR/MRP), member 3 transporter ABCG1 ATP-binding cassette, sub-family G (WHITE), member 1 transporter ABCG8 ATP-binding cassette, sub-family G (WHITE), member 8 transporter ACE angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 peptidase ACTR2 ARP2 actin-related protein 2 homolog (yeast) other ACVR1 activin A receptor, type I kinase ACVRL1 activin A receptor type II-like 1 kinase ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) peptidase ADAM10 ADAM metallopeptidase domain 10 peptidase ADAM17 ADAM metallopeptidase domain 17 peptidase ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) peptidase ADAM22 ADAM metallopeptidase domain 22 peptidase ADAM21 ADAM metallopeptidase domain 21 peptidase ADCY1 adenylate cyclase 1 (brain) enzyme ADCY3 adenylate cyclase 3 enzyme ADCY5 adenylate cyclase 5 enzyme aminoacyl tRNA synthetase complex-interacting multifunctional AIMP2 protein 2 other ALK anaplastic lymphoma receptor tyrosine kinase kinase ANXA1 annexin A1 other ANXA2 annexin A2 other ANXA4 annexin A4 other ANXA5 annexin A5 other ANXA6 -
Cytokine Priming of Naïve CD8+ T Lymphocytes Modulates Chromatin
bioRxiv preprint doi: https://doi.org/10.1101/2020.08.11.246553; this version posted August 12, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Cytokine priming of naïve CD8+ T lymphocytes modulates chromatin accessibility that partially overlaps with changes induced by antigen simulation Akouavi Julite Quenum1, Maryse Cloutier2, Madanraj Appiya Santharam1, Marian Mayhue1, Sheela Ramanathan1,2,3 and Subburaj Ilangumaran1,2,3 1 Cell biology graduate program, 2 Immunology graduate program, Department of Immunology and Cell Biology, Faculty of Medicine and Health Sciences, Université de Sherbrooke. 3 CRCHUS, Sherbrooke, Québec J1H 5N4, Canada Corresponding author: SubburaJ Ilangumaran, Ph.D. Department of Immunology and Cell Biology Faculty of Medicine and Health Sciences Université de Sherbrooke 3001 North 12th avenue, Sherbrooke QC J1H 5N4, Canada Tel: 1-819-346 1110 x14834; Fax: 1-819-564 5215 [email protected] Running title: ATACseq of cytokine-primed CD8+ T cells Keywords: CD8+ T cells, IL-15, IL-21, TCR, cytokine priming, ATACseq. 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.08.11.246553; this version posted August 12, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Abstract: 247 words Illustrations: Colour Figures: 6 Tables: 2 Supplementary Figures: 5 Supplementary Tables: 8 2 bioRxiv preprint doi: https://doi.org/10.1101/2020.08.11.246553; this version posted August 12, 2020.