Appendix 1 Appendix 2
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
APP203875 Submissions Compilation.Pdf
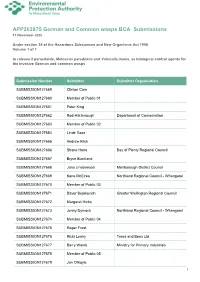
APP203875 German and Common wasps BCA Submissions 11 November 2020 Under section 34 of the Hazardous Substances and New Organisms Act 1996 Volume 1 of 1 to release 2 parasitoids, Metoecus paradoxus and Volucella inanis, as biological control agents for the invasive German and common wasps Submission Number Submitter Submitter Organisation SUBMISSION127659 Clinton Care SUBMISSION127660 Member of Public 01 SUBMISSION127661 Peter King SUBMISSION127662 Rod Hitchmough Department of Conservation SUBMISSION127663 Member of Public 02 SUBMISSION127664 Linde Rose SUBMISSION127665 Andrew Blick SUBMISSION127666 Shane Hona Bay of Plenty Regional Council SUBMISSION127667 Bryce Buckland SUBMISSION127668 Jono Underwood Marlborough District Council SUBMISSION127669 Kane McElrea Northland Regional Council - Whangarei SUBMISSION127670 Member of Public 03 SUBMISSION127671 Davor Bejakovich Greater Wellington Regional Council SUBMISSION127672 Margaret Hicks SUBMISSION127673 Jenny Dymock Northland Regional Council - Whangarei SUBMISSION127674 Member of Public 04 SUBMISSION127675 Roger Frost SUBMISSION127676 Ricki Leahy Trees and Bees Ltd SUBMISSION127677 Barry Wards Ministry for Primary Industries SUBMISSION127678 Member of Public 05 SUBMISSION127679 Jan O'Boyle 1 Submission Number Submitter Submitter Organisation Apiculture New Zealand Science and SUBMISSION127680 Sue Carter Research Focus Group SUBMISSION127681 Andrea Dorn SUBMISSION127682 Benita Wakefield Te Rūnanga o Ngāi Tahu SUBMISSION127683 Emma Edney-Browne Auckland Council SUBMISSION127684 David Hunter -

White Admiral Newsletter
W h i t e A d m i r a l Newsletter 88 Summer 2014 Suffolk Naturalists’ Society C o n te n t s E d i t or i a l Ben Heather 1 Another new fungus for Suffolk Neil Mahler 2 The battle continues…the fight Matt Holden 4 against invasive alien plants in the Stour Valley! Nesting Materials Richard Stewart 8 How you can help monitor Suffolk’s Su e H o ot on 8 b a t s ? Records please! Rosemary Leaf Beetle Ben Heather 10 Stratiomys longicornis – a fly a long Peter Vincent 11 way from home! Periglacial Landforms in Breckland Caroline Markham 13 Are some roadside plants on the Dr. Anne Kell and 15 verge of extinction? Dennis Kell Where has all the road kill gone? Tom Langton 21 Back on the Hopper Trail in 2013 Colin Lucas & 22 Tricia Taylor Volucella zonaria – an impressive Peter Vincent 24 b e a s t Suffolk Show Wildlife H a w k H on e y 27 Suffolk’s Nature Strategy Nick Collinson 29 Species ‘Re - introductions’ Nick Miller 32 ISSN 0959-8537 Published by the Suffolk Naturalists’ Society c/o Ipswich Museum, High Street, Ipswich, Suffolk IP1 3QH Registered Charity No. 206084 © Suffolk Naturalists’ Society Front cover: Alder spittle bug - Aphrophora alni by Ben Heather Newsletter 88 - Summer 2014 Thank you to all those who have contributed to this full issue of the White Admiral newsletter. This issue covers a wide range of topics from roadside verges to an observation on the lack of roadkill on our roads. -

Read the Decision
Decision Date 5 February 2021 Application number APP203875 Application type To import for release and/or release from containment any new organism under section 34 of the Hazardous Substances and New Organisms Act 1996 Applicant Tasman District Council Date of hearing/consideration 17 December 2020 Date Application received 14 September 2020 Considered by A decision-making committee of the Environmental Protection Authority (the Committee)1 Dr Nick Roskruge (Chair) Dr John Taylor Mr Peter Cressey Purpose of the application To import and release two parasitoids, Metoecus paradoxus and Volucella inanis as biological control agents for the invasive German and common wasps (Vespula germanica and V. vulgaris). New organism approved Metoecus paradoxus Linnaeus 1761 Volucella inanis Linnaeus 1758 1 The Committee referred to in this decision is the subcommittee that has made the decision on this application under delegated authority in accordance with section 18A of the Act. Decision APP203875 Summary of decision 1. Application APP203875 to import and release two parasitoids, Metoecus paradoxus and Volucella inanis, as biological control agents (BCAs) for the invasive German and common wasps (Vespula germanica and V. vulgaris), in New Zealand, was lodged under section 34 of the Hazardous Substances and New Organisms Act 1996 (the Act). 2. The application was considered in accordance with the relevant provisions of the Act and of the HSNO (Methodology) Order 1998 (the Methodology). 3. The Committee has approved the application in accordance with section 38(1)(a) of the Act. Application and consideration process 4. The application was formally received on 14 September 2020. 5. The applicant, Tasman District Council, applied to the Environmental Protection Authority (EPA) to import and release two parasitoids, Metoecus paradoxus and Volucella inanis, as BCAs for the invasive German and common wasps (Vespula germanica and V. -

HOVERFLY NEWSLETTER Dipterists
HOVERFLY NUMBER 41 NEWSLETTER SPRING 2006 Dipterists Forum ISSN 1358-5029 As a new season begins, no doubt we are all hoping for a more productive recording year than we have had in the last three or so. Despite the frustration of recent seasons it is clear that national and international study of hoverflies is in good health, as witnessed by the success of the Leiden symposium and the Recording Scheme’s report (though the conundrum of the decline in UK records of difficult species is mystifying). New readers may wonder why the list of literature references from page 15 onwards covers publications for the year 2000 only. The reason for this is that for several issues nobody was available to compile these lists. Roger Morris kindly agreed to take on this task and to catch up for the missing years. Each newsletter for the present will include a list covering one complete year of the backlog, and since there are two newsletters per year the backlog will gradually be eliminated. Once again I thank all contributors and I welcome articles for future newsletters; these may be sent as email attachments, typed hard copy, manuscript or even dictated by phone, if you wish. Please do not forget the “Interesting Recent Records” feature, which is rather sparse in this issue. Copy for Hoverfly Newsletter No. 42 (which is expected to be issued with the Autumn 2006 Dipterists Forum Bulletin) should be sent to me: David Iliff, Green Willows, Station Road, Woodmancote, Cheltenham, Glos, GL52 9HN, (telephone 01242 674398), email: [email protected], to reach me by 20 June 2006. -

Hoverflies Family: Syrphidae
Birmingham & Black Country SPECIES ATLAS SERIES Hoverflies Family: Syrphidae Andy Slater Produced by EcoRecord Introduction Hoverflies are members of the Syrphidae family in the very large insect order Diptera ('true flies'). There are around 283 species of hoverfly found in the British Isles, and 176 of these have been recorded in Birmingham and the Black Country. This atlas contains tetrad maps of all of the species recorded in our area based on records held on the EcoRecord database. The records cover the period up to the end of 2019. Myathropa florea Cover image: Chrysotoxum festivum All illustrations and photos by Andy Slater All maps contain Contains Ordnance Survey data © Crown Copyright and database right 2020 Hoverflies Hoverflies are amongst the most colourful and charismatic insects that you might spot in your garden. They truly can be considered the gardener’s fiend as not only are they important pollinators but the larva of many species also help to control aphids! Great places to spot hoverflies are in flowery meadows on flowers such as knapweed, buttercup, hogweed or yarrow or in gardens on plants such as Canadian goldenrod, hebe or buddleia. Quite a few species are instantly recognisable while the appearance of some other species might make you doubt that it is even a hoverfly… Mimicry Many hoverfly species are excellent mimics of bees and wasps, imitating not only their colouring, but also often their shape and behaviour. Sometimes they do this to fool the bees and wasps so they can enter their nests to lay their eggs. Most species however are probably trying to fool potential predators into thinking that they are a hazardous species with a sting or foul taste, even though they are in fact harmless and perfectly edible. -

Encyclopedia of Social Insects
G Guests of Social Insects resources and homeostatic conditions. At the same time, successful adaptation to the inner envi- Thomas Parmentier ronment shields them from many predators that Terrestrial Ecology Unit (TEREC), Department of cannot penetrate this hostile space. Social insect Biology, Ghent University, Ghent, Belgium associates are generally known as their guests Laboratory of Socioecology and Socioevolution, or inquilines (Lat. inquilinus: tenant, lodger). KU Leuven, Leuven, Belgium Most such guests live permanently in the host’s Research Unit of Environmental and nest, while some also spend a part of their life Evolutionary Biology, Namur Institute of cycle outside of it. Guests are typically arthropods Complex Systems, and Institute of Life, Earth, associated with one of the four groups of eusocial and the Environment, University of Namur, insects. They are referred to as myrmecophiles Namur, Belgium or ant guests, termitophiles, melittophiles or bee guests, and sphecophiles or wasp guests. The term “myrmecophile” can also be used in a broad sense Synonyms to characterize any organism that depends on ants, including some bacteria, fungi, plants, aphids, Inquilines; Myrmecophiles; Nest parasites; and even birds. It is used here in the narrow Symbionts; Termitophiles sense of arthropods that associated closely with ant nests. Social insect nests may also be parasit- Social insect nests provide a rich microhabitat, ized by other social insects, commonly known as often lavishly endowed with long-lasting social parasites. Although some strategies (mainly resources, such as brood, retrieved or cultivated chemical deception) are similar, the guests of food, and nutrient-rich refuse. Moreover, nest social insects and social parasites greatly differ temperature and humidity are often strictly regu- in terms of their biology, host interaction, host lated. -

Diptera, Sy Ae)
Ce nt re fo r Eco logy & Hydrology N AT U RA L ENVIRO N M EN T RESEA RC H CO U N C IL Provisional atlas of British hover les (Diptera, Sy ae) _ Stuart G Ball & Roger K A Morris _ J O I N T NATURE CONSERVATION COMMITTEE NERC Co pyright 2000 Printed in 2000 by CRL Digital Limited ISBN I 870393 54 6 The Centre for Eco logy an d Hydrolo gy (CEI-0 is one of the Centres an d Surveys of the Natu ral Environme nt Research Council (NERC). Established in 1994, CEH is a multi-disciplinary , environmental research organisation w ith som e 600 staff an d w ell-equipp ed labo ratories and field facilities at n ine sites throughout the United Kingdom . Up u ntil Ap ril 2000, CEM co m prise d of fou r comp o nent NERC Institutes - the Institute of Hydrology (IH), the Institute of Freshw ater Eco logy (WE), the Institute of Terrestrial Eco logy (ITE), and the Institute of Virology an d Environmental Micro b iology (IVEM). From the beginning of Ap dl 2000, CEH has operated as a single institute, and the ind ividual Institute nam es have ceased to be used . CEH's mission is to "advance th e science of ecology, env ironme ntal microbiology and hyd rology th rough h igh q uality and inte rnat ionall) recognised research lead ing to better understanding and quantifia ttion of the p hysical, chem ical and b iolo gical p rocesses relating to land an d freshwater an d living organisms within the se environments". -

An Application to Import and Release Two Parasitoids to Control German and Common Wasps
APP203875: An application to import and release two parasitoids to control German and common wasps. December 2020 The application Tasman District Council lodged an application with the EPA on 14 September 2020 seeking approval to release Metoecus paradoxus and Volucella inanis, as biological control agents for the social wasp, Vespula germanica and V. Vulgaris. The application was publicly notified: - 25 support, - 2 neither supported nor opposed, and - 3 opposed the application. 2 The biocontrol agents Metoecus paradoxus Volucella inanis Wasp-nest beetle Hoverfly Photo by B. Brown Photo by B. Brown . Target mainly Vespula vulgaris . Target species in the subfamilies Vespinae . Adults short lived and do not feed . Adults feed on pollen . Female lays several hundred eggs . Female lays 300-660 eggs . 1 wasp larva per beetle . 2 wasp larvae per hoverfly . Lack of host selection from the larvae 3 The target hosts Vespula vulgaris Vespula germanica Common wasp German wasp • Accidentally introduced • Widespread and thrive in New Zealand 4 • Highest concentration of social wasp Host specificity Host range testing . Volucella inanis does not target bumblebees Phylogeny Order Suborder Infraorder Superfamily Parasitica Chrysidoidea (cuckoo wasps and allies) ‘Parasitic wasps’ Vespoidea (potter, paper, and other wasps) Symphyta Sierolomorphoidea Tiphioidea Aculeata Hymenoptera Apocrita Thynnoidea ‘Stinging wasps’ Pompiloidea (spider wasps) Scolioidea (scoliid wasps and allies) Formicoidea (ants) 5 Apoidea (speciform wasps, bumblebees and bees) Host specificity Behaviour: social versus solitary . No native social bees or wasps (except native ants) . Valued exotic social species not targeted Life cycle . Similar to honeybees and bumblebees . Different to native solitary species . Obstacles for the BCAs: - Small size - Nest entrance size - No comb Photo by J. -

Does Phenology Explain Plant-Pollinator Interactions at Different Latitudes? an Assessment of Its
1 Title: Does phenology explain plant-pollinator interactions at different latitudes? An assessment of its 2 explanatory power in plant-hoverfly networks in French calcareous grasslands 3 Authors: Natasha de Manincor¹*, Nina Hautekeete¹, Yves Piquot¹, Bertrand Schatz², Cédric 4 Vanappelghem³, François Massol¹,4 5 ¹Université de Lille, CNRS, UMR 8198 - Evo-Eco-Paleo, F-59000 Lille, France 6 ²CEFE, EPHE-PSL, CNRS, University of Montpellier, University of Paul Valéry Montpellier 3, IRD, 7 Montpellier, France 8 ³Conservatoire d'espaces naturels Nord et du Pas-de-Calais, 160 rue Achille Fanien - ZA de la Haye, 9 62190 LILLERS 10 4Univ. Lille, CNRS, Inserm, CHU Lille, Institut Pasteur de Lille, U1019 - UMR 8204 - CIIL - Center for 11 Infection and Immunity of Lille, F-59000 Lille, France 12 13 E-mail addresses and ORCID numbers: 14 Natasha de Manincor: [email protected], [email protected], 0000- 15 0001-9696-125X 16 Nina Hautekeete: [email protected], 0000-0002-6071-5601 17 Yves Piquot: [email protected], 0000-0001-9977-8936 18 Bertrand Schatz: [email protected], 0000-0003-0135-8154 19 Cédric Vanappelghem: [email protected] 20 François Massol: [email protected], 0000-0002-4098-955X 21 22 Short title: Phenology and plant-hoverfly interactions 23 Keywords: Bayesian model, interaction probability, latent block model, latitudinal gradient, 24 mutualistic network, phenology overlap, species abundance, structural equation model. 1 25 *Corresponding author information: Natasha de Manincor, e-mail: [email protected], 26 [email protected], phone: +330362268530 27 Author contributions 28 NDM and FM conceived the project, formulated and implemented the model. -

Diptera: Tachinidae) James Lumbers, B
Keynote Address Making taxonomy and collections-based research relevant in a changing world Bryan Lessard Australian National Insect Collection, CSIRO, Black Mountain, Clunies Ross Street, Acton, ACT, 2601 In a time of taxonomic decline, insect apocalypses and fake news, it’s becoming increasingly more important to conduct and promote our biodiversity research. Recent advances in technology have breathed new life into natural history collections: metabarcoding allows us to identify new insect pollinators in threatened alpine zones; whole genome shotgun sequencing allows us to map mitochondrial genomes from pinned specimens of medially important mosquitoes; and transcriptome sequencing allow us to identify protein-coding genes that cause anaphylactic reactions to horse fly bites. Modern digitisation has also rapidly increased the speed at which we database our collections, opening them to the public and other researchers throughout the world. Taxonomic knowledge and collection data can be useful to inform industry and government policy, as a recent taxonomic revision of the Australian soldier flies led to collaborations with small to medium sized businesses and the opportunity to advise policy makers on the use of insects as livestock feed. Collections can also provide specimen data to support global industry trends, as the United Nations estimate that the emerging edible insect market will be worth $1.5 billion by 2023. A recent Australian edible insect symposium identified a commercial need for native edible insect species that are currently being identified using entomological collections. New platforms of communication like social media have revolutionised the way we share our science in real time and allow us to reach a wider audience formed of the general public, researchers and industry. -

Download Vol. 6 (Autumn 2012)
Shropshire Entomology – October 2012 (No.6) A bi-annual newsletter focussing upon the study of insects and other invertebrates in the county of Shropshire (V.C. 40) October 2012 (Vol. 6) Editor: Pete Boardman [email protected] ~ Welcome ~ Welco me to the 6th edition of the Shropshire Entomology newsletter. I hope you enjoy it and it inspires you to submit your own articles relating to any aspect of entomology relevant to Shropshire or Shropshire entomologists. Many thanks once more to everyone who has contributed to this edition. The deadline for submission of content for Vol. 7 is Friday 8th March 2013. Please feel free to pass this newsletter on to anyone you feel might be interested in it. Please note there are instructions for authors at the back of this newsletter. Note – past newsletters are now available for download as PDF’s from www.invertebrate-challenge.org.uk/newsletters- and -resources.aspx ~ Contents ~ Identification of a Shropshire pseudoscorpion – From the rarest to the most common!: Dr Kris Hart Wood ants in the Wyre Forest: Paul Wilson Your last chance.....: Sue McLamb Dudmaston’s rare and interesting insects: Caroline Uff Dudmaston, Shropshire’s top aculeate site?: Ian Cheeseborough The plant bug Orsillus depressus (Mulsant & Rey, 1852) new to Shropshire: Maria Justamond Not a total washout! The red-veined darter Sympetrum fonscolombii Selys, 1840 in Shropshire: Sue McLamb Granville Country Park butterfly count: Keith Fowler An encounter with Stylops: Maria Justamond Potential identification problems with Poeculis (Coleoptera: Carabidae): Michelle Furber & Warren Putter Trampolining beetles: Caroline Uff New and ‘nearly new’ craneflies to Shropshire during 2012: Pete Boardman Insects are your friends – an occasional guide to insect identification by reference to unexpected sources – No.1 – The Dame Edna fly Palloptera mulleibris (Harris [1780]): C. -

Neue Schwebfliegen-Literatur
ZOBODAT - www.zobodat.at Zoologisch-Botanische Datenbank/Zoological-Botanical Database Digitale Literatur/Digital Literature Zeitschrift/Journal: Volucella - Die Schwebfliegen-Zeitschrift Jahr/Year: 2004 Band/Volume: 7 Autor(en)/Author(s): Dziock Franz, Schmid Ulrich Artikel/Article: Neue Schwebfliegen-Literatur (7) 223-237 ©Volucella; Dieter Doczkal (München) und Ulrich Schmid (Stuttgart), download www.zobodat.at 222 VOLUCELLA 7, 2004 Dziock & Schmid: Neue Schwebfliegen-Literatur (7) 223 Neue Schwebfliegen-Literatur (7) Frank Dziock und Ulrich Schmid Dziock, F.; Schmid, U. (2004): New literature on hoverflies (Diptera, Syrphidae) (7). – Volucella 7, 223-237. Stuttgart. Achterkamp, B. (2002): Episyrphusprojekt her- Applicata 108(2), 115-124. nieuws in 2002. – Zweefvliegennieuwsbrief Baéz, M.; Barkemeyer, W. (2002): The identity of 6(1), 28-29. Eumerus terminalis Santos Abreu, 1924. – Stu- Achterkamp, B. (2002): Tussenstand Episyrphuspro- dia dipterologica 9, 518. jekt de maand mei. – Zweefvliegennieuwsbrief Ball, S.; Morris, R. (2003): News from the Hoverfly 6(2), 6-7. Recording Scheme. – Hoverfly Newsletter 36, Agarwala, B. K.; Bardhanroy, P.; Yasuda, H.; 9-11. Takizawa, T. (2003): Effects of conspecific Barkalov, A. V. (2003): Description of the new Palae- and heterospecific competitors on feeding and arctic species Volucella bella (Diptera, Syrphi- oviposition of a predatory ladybird: a laboratory dae) from the zonaria group. – Zoologichesky study. – Entomologia Experimentalis et Appli- Zhurnal 82, 1133-1137. cata 106, 219-226. Barkalov, A.V. (2002): A subgeneric classification Anderson, A.J.; McOwan, P.W. (2003): Model of a of the genus Cheilosia Meigen, 1822 (Diptera, predatory stealth behaviour camouflaging moti- Syrphidae). – Entomologicheskoe Obozrenie on. – Proceedings of the Royal Society Biologi- 81(1), 218-234, 261.