For Review Only
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Biolphilately Vol-64 No-3
BIOPHILATELY OFFICIAL JOURNAL OF THE BIOLOGY UNIT OF ATA MARCH 2020 VOLUME 69, NUMBER 1 Great fleas have little fleas upon their backs to bite 'em, And little fleas have lesser fleas, and so ad infinitum. —Augustus De Morgan Dr. Indraneil Das Pangolins on Stamps More Inside >> IN THIS ISSUE NEW ISSUES: ARTICLES & ILLUSTRATIONS: From the Editor’s Desk ......................... 1 Botany – Christopher E. Dahle ............ 17 Pangolins on Stamps of the President’s Message .............................. 2 Fungi – Paul A. Mistretta .................... 28 World – Dr. Indraneil Das ..................7 Secretary -Treasurer’s Corner ................ 3 Mammalia – Michael Prince ................ 31 Squeaky Curtain – Frank Jacobs .......... 15 New Members ....................................... 3 Ornithology – Glenn G. Mertz ............. 35 New Plants in the Philatelic News of Note ......................................... 3 Ichthyology – J. Dale Shively .............. 57 Herbarium – Christopher Dahle ....... 23 Women’s Suffrage – Dawn Hamman .... 4 Entomology – Donald Wright, Jr. ........ 59 Rats! ..................................................... 34 Event Calendar ...................................... 6 Paleontology – Michael Kogan ........... 65 New Birds in the Philatelic Wedding Set ........................................ 16 Aviary – Charles E. Braun ............... 51 Glossary ............................................... 72 Biology Reference Websites ................ 69 ii Biophilately March 2020 Vol. 69 (1) BIOPHILATELY BIOLOGY UNIT -

Changes in Arthropod Abundance and Diversity with Invasive
CHANGES IN ARTHROPOD ABUNDANCE AND DIVERSITY WITH INVASIVE GRASSES A Thesis by ERIN E. CORD Submitted to the College of Graduate Studies Texas A&M University-Kingsville in partial fulfillment of the requirements for the degree of MASTER OF SCIENCE August 2011 Major Subject: Range and Wildlife Management CHANGES IN ARTHROPOD ABUNDANCE AND DIVERSITY WITH INVASIVE GRASSES A Thesis by ERIN E. CORD Approved as to style and content by: ______________________________ Andrea R. Litt, Ph.D. (Chairman of Committee) ___________________________ ___________________________ Timothy E. Fulbright, Ph.D. Greta L. Schuster, Ph.D. (Member) (Member) _____________________________ Scott E. Henke, Ph.D. (Chair of Department) _________________________________ Ambrose Anoruo, Ph.D. (Associate VP for Research & Dean, College of Graduate Studies) August 2011 ABSTRACT Changes in Arthropod Abundance and Diversity with Invasive Grasses (August 2011) Erin E. Cord, B.S., University Of Delaware Chairman of Committee: Dr. Andrea R. Litt Invasive grasses can alter plant communities and can potentially affect arthropods due to specialized relationships with certain plants as food resources and reproduction sites. Kleberg bluestem (Dichanthium annulatum) is a non-native grass and tanglehead (Heteropogon contortus) is native to the United States, but recently has become dominant in south Texas. I sought to: 1) quantify changes in plant and arthropod communities in invasive grasses compared to native grasses, and 2) determine if grass origin would alter effects. I sampled vegetation and arthropods on 90 grass patches in July and September 2009 and 2010 on the King Ranch in southern Texas. Arthropod communities in invasive grasses were less diverse and abundant, compared to native grasses; I also documented differences in presence and abundance of certain orders and families. -

Final Report for the University of Nottingham / Operation Wallacea Forest Projects, Honduras 2004
FINAL REPORT for the University of Nottingham / Operation Wallacea forest projects, Honduras 2004 TABLE OF CONTENTS FINAL REPORT FOR THE UNIVERSITY OF NOTTINGHAM / OPERATION WALLACEA FOREST PROJECTS, HONDURAS 2004 .....................................................................................................................................................1 INTRODUCTION AND OVERVIEW ..............................................................................................................................3 List of the projects undertaken in 2004, with scientists’ names .........................................................................4 Forest structure and composition ..................................................................................................................................... 4 Bat diversity and abundance ............................................................................................................................................ 4 Bird diversity, abundance and ecology ............................................................................................................................ 4 Herpetofaunal diversity, abundance and ecology............................................................................................................. 4 Invertebrate diversity, abundance and ecology ................................................................................................................ 4 Primate behaviour........................................................................................................................................................... -

A Distributional Study of the Butterflies of the Sierra De Tuxtla in Veracruz, Mexico. Gary Noel Ross Louisiana State University and Agricultural & Mechanical College
Louisiana State University LSU Digital Commons LSU Historical Dissertations and Theses Graduate School 1967 A Distributional Study of the Butterflies of the Sierra De Tuxtla in Veracruz, Mexico. Gary Noel Ross Louisiana State University and Agricultural & Mechanical College Follow this and additional works at: https://digitalcommons.lsu.edu/gradschool_disstheses Recommended Citation Ross, Gary Noel, "A Distributional Study of the Butterflies of the Sierra De Tuxtla in Veracruz, Mexico." (1967). LSU Historical Dissertations and Theses. 1315. https://digitalcommons.lsu.edu/gradschool_disstheses/1315 This Dissertation is brought to you for free and open access by the Graduate School at LSU Digital Commons. It has been accepted for inclusion in LSU Historical Dissertations and Theses by an authorized administrator of LSU Digital Commons. For more information, please contact [email protected]. This dissertation has been microfilmed exactly as received 67-14,010 ROSS, Gary Noel, 1940- A DISTRIBUTIONAL STUDY OF THE BUTTERFLIES OF THE SIERRA DE TUXTLA IN VERACRUZ, MEXICO. Louisiana State University and Agricultural and Mechanical CoUege, Ph.D., 1967 Entomology University Microfilms, Inc., Ann Arbor, Michigan A DISTRIBUTIONAL STUDY OF THE BUTTERFLIES OF THE SIERRA DE TUXTLA IN VERACRUZ, MEXICO A D issertation Submitted to the Graduate Faculty of the Louisiana State University and A gricultural and Mechanical College in partial fulfillment of the requirements for the degree of Doctor of Philosophy in The Department of Entomology by Gary Noel Ross M.S., Louisiana State University, 196*+ May, 1967 FRONTISPIECE Section of the south wall of the crater of Volcan Santa Marta. May 1965, 5,100 feet. ACKNOWLEDGMENTS Many persons have contributed to and assisted me in the prep aration of this dissertation and I wish to express my sincerest ap preciation to them all. -

Uehara-Prado Marcio D.Pdf
FICHA CATALOGRÁFICA ELABORADA PELA BIBLIOTECA DO INSTITUTO DE BIOLOGIA – UNICAMP Uehara-Prado, Marcio Ue3a Artrópodes terrestres como indicadores biológicos de perturbação antrópica / Marcio Uehara do Prado. – Campinas, SP: [s.n.], 2009. Orientador: André Victor Lucci Freitas. Tese (doutorado) – Universidade Estadual de Campinas, Instituto de Biologia. 1. Indicadores (Biologia). 2. Borboleta . 3. Artrópode epigéico. 4. Mata Atlântica. 5. Cerrados. I. Freitas, André Victor Lucci. II. Universidade Estadual de Campinas. Instituto de Biologia. III. Título. (rcdt/ib) Título em inglês: Terrestrial arthropods as biological indicators of anthropogenic disturbance. Palavras-chave em inglês : Indicators (Biology); Butterflies; Epigaeic arthropod; Mata Atlântica (Brazil); Cerrados. Área de concentração: Ecologia. Titulação: Doutor em Ecologia. Banca examinadora: André Victor Lucci Freitas, Fabio de Oliveira Roque, Paulo Roberto Guimarães Junior, Flavio Antonio Maës dos Santos, Thomas Michael Lewinsohn. Data da defesa : 21/08/2009. Programa de Pós-Graduação: Ecologia. iv Dedico este trabalho ao professor Keith S. Brown Jr. v AGRADECIMENTOS Ao longo dos vários anos da tese, muitas pessoas contribuiram direta ou indiretamente para a sua execução. Gostaria de agradecer nominalmente a todos, mas o espaço e a memória, ambos limitados, não permitem. Fica aqui o meu obrigado geral a todos que me ajudaram de alguma forma. Ao professor André V.L. Freitas, por sempre me incentivar e me apoiar em todos os momentos da tese, e por todo o ensinamento passado ao longo de nossa convivência de uma década. A minha família: Dona Júlia, Bagi e Bete, pelo apoio incondicional. A Cris, por ser essa companheira incrível, sempre cuidando muito bem de mim. A todas as meninas que participaram do projeto original “Artrópodes como indicadores biológicos de perturbação antrópica em Floresta Atlântica”, em especial a Juliana de Oliveira Fernandes, Huang Shi Fang, Mariana Juventina Magrini, Cristiane Matavelli, Tatiane Gisele Alves e Regiane Moreira de Oliveira. -

INSECTA: LEPIDOPTERA) DE GUATEMALA CON UNA RESEÑA HISTÓRICA Towards a Synthesis of the Papilionoidea (Insecta: Lepidoptera) from Guatemala with a Historical Sketch
ZOOLOGÍA-TAXONOMÍA www.unal.edu.co/icn/publicaciones/caldasia.htm Caldasia 31(2):407-440. 2009 HACIA UNA SÍNTESIS DE LOS PAPILIONOIDEA (INSECTA: LEPIDOPTERA) DE GUATEMALA CON UNA RESEÑA HISTÓRICA Towards a synthesis of the Papilionoidea (Insecta: Lepidoptera) from Guatemala with a historical sketch JOSÉ LUIS SALINAS-GUTIÉRREZ El Colegio de la Frontera Sur (ECOSUR). Unidad Chetumal. Av. Centenario km. 5.5, A. P. 424, C. P. 77900. Chetumal, Quintana Roo, México, México. [email protected] CLAUDIO MÉNDEZ Escuela de Biología, Universidad de San Carlos, Ciudad Universitaria, Campus Central USAC, Zona 12. Guatemala, Guatemala. [email protected] MERCEDES BARRIOS Centro de Estudios Conservacionistas (CECON), Universidad de San Carlos, Avenida La Reforma 0-53, Zona 10, Guatemala, Guatemala. [email protected] CARMEN POZO El Colegio de la Frontera Sur (ECOSUR). Unidad Chetumal. Av. Centenario km. 5.5, A. P. 424, C. P. 77900. Chetumal, Quintana Roo, México, México. [email protected] JORGE LLORENTE-BOUSQUETS Museo de Zoología, Facultad de Ciencias, UNAM. Apartado Postal 70-399, México D.F. 04510; México. [email protected]. Autor responsable. RESUMEN La riqueza biológica de Mesoamérica es enorme. Dentro de esta gran área geográfi ca se encuentran algunos de los ecosistemas más diversos del planeta (selvas tropicales), así como varios de los principales centros de endemismo en el mundo (bosques nublados). Países como Guatemala, en esta gran área biogeográfi ca, tiene grandes zonas de bosque húmedo tropical y bosque mesófi lo, por esta razón es muy importante para analizar la diversidad en la región. Lamentablemente, la fauna de mariposas de Guatemala es poco conocida y por lo tanto, es necesario llevar a cabo un estudio y análisis de la composición y la diversidad de las mariposas (Lepidoptera: Papilionoidea) en Guatemala. -

Aggregate?Pinheiro Et Al
610 Why do the ithomiines (Lepidoptera, Nymphalidae) aggregate?Pinheiro et al. Notes on a butterfly pocket in central Brazil 1 2 3 Carlos E. G. Pinheiro , Ísis Meri Medri & Ana Karina Moreyra Salcedo 1Departamento de Zoologia, Universidade de Brasília – UnB, 70910-900 Brasília-DF, Brasil. [email protected] 2,3Programa de Pós-graduação em Ecologia, Depto. de Ecologia, Universidade de Brasília – UnB, 70910-900 Brasília-DF, Brasil. [email protected] ABSTRACT. Why do the ithomiines (Lepidoptera, Nymphalidae) aggregate? Notes on a butterfly pocket in central Brazil. This study provides information on the species composition and the number of butterflies in different phases of an ithomiine aggregation during the 2004 dry season in central Brazil, and tests some hypotheses concerning the pocket formation. The results obtained suggest that ithomiine pockets constitute primarily an adaptation of butterflies to the adverse climatic conditions of the dry season, such as high temperatures and low air relative humidity, rather than the occurrence of large concentrations of adult food resources (flowers visited for nectar were not found in the pocket site) or defense against visually hunting predators (contrary to the prediction tested, the frequency of butterflies bearing birds beak marks on the wings significantly increased along the period of pocket formation, especially in the case of Mechanitis polymnia, the most abundant species in the pocket). Other hypotheses concerning the pocket formation are also discussed. KEYWORDS. Beak marks; insectivorous birds; ithomiine pockets; Müllerian mimicry. RESUMO. Por que os Ithomiinae (Lepidoptera, Nymphalidae) se agregam? Observações sobre um bolsão de borboletas no Brasil central. Este trabalho apresenta dados sobre a composição de espécies e o número de indivíduos encontrados em diferentes fases de formação de um bolsão de Ithomiinae investigado na estação seca de 2004 em uma floresta de galeria do Brasil central, e testa algumas hipóteses relacionadas à formação do bolsão. -

Developmental, Cellular, and Biochemical
Developmental, cellular, and biochemical basis of transparency in the glasswing butterfly Greta oto Aaron Pomerantz, Radwanul Siddique, Elizabeth Cash, Yuriko Kishi, Charline Pinna, Kasia Hammar, Doris Gomez, Marianne Elias, Nipam Patel To cite this version: Aaron Pomerantz, Radwanul Siddique, Elizabeth Cash, Yuriko Kishi, Charline Pinna, et al.. Devel- opmental, cellular, and biochemical basis of transparency in the glasswing butterfly Greta oto. 2020. hal-03012452 HAL Id: hal-03012452 https://hal.archives-ouvertes.fr/hal-03012452 Preprint submitted on 18 Nov 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. bioRxiv preprint doi: https://doi.org/10.1101/2020.07.02.183590; this version posted July 2, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Title 2 Developmental, cellular, and biochemical basis of transparency in the glasswing butterfly 3 Greta oto 4 5 Authors 6 Aaron F. Pomerantz1,2*, Radwanul H. Siddique3,4, Elizabeth I. Cash5, Yuriko Kishi6,7, 7 Charline Pinna8, Kasia Hammar2, Doris Gomez9, Marianne Elias8, Nipam H. -

The Brain of a Nocturnal Migratory Insect, the Australian Bogong Moth
bioRxiv preprint doi: https://doi.org/10.1101/810895; this version posted January 21, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. The brain of a nocturnal migratory insect, the Australian Bogong moth Authors: Andrea Adden1, Sara Wibrand1, Keram Pfeiffer2, Eric Warrant1, Stanley Heinze1,3 1 Lund Vision Group, Lund University, Sweden 2 University of Würzburg, Germany 3 NanoLund, Lund University, Sweden Correspondence: [email protected] Abstract Every year, millions of Australian Bogong moths (Agrotis infusa) complete an astonishing journey: in spring, they migrate over 1000 km from their breeding grounds to the alpine regions of the Snowy Mountains, where they endure the hot summer in the cool climate of alpine caves. In autumn, the moths return to their breeding grounds, where they mate, lay eggs and die. These moths can use visual cues in combination with the geomagnetic field to guide their flight, but how these cues are processed and integrated in the brain to drive migratory behavior is unknown. To generate an access point for functional studies, we provide a detailed description of the Bogong moth’s brain. Based on immunohistochemical stainings against synapsin and serotonin (5HT), we describe the overall layout as well as the fine structure of all major neuropils, including the regions that have previously been implicated in compass-based navigation. The resulting average brain atlas consists of 3D reconstructions of 25 separate neuropils, comprising the most detailed account of a moth brain to date. -
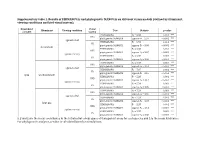
Supplementary Table 1. Results of Permanovas and Phylogenetic Manovas on Different Vision Models (Defined by Illuminant, Viewing Conditions and Bird Visual System)
Supplementary table 1. Results of PERMANOVAs and phylogenetic MANOVAs on different vision models (defined by illuminant, viewing conditions and bird visual system). Dependent Visual Illuminant Viewing condition Test Statistic p-value variable system PERMANOVA F9 = 6.88 0.001 *** UVS phylogenetic MANOVA approx-F9 = 2.97 < 0.001 *** against a leaf PERMANOVA F9 = 6.93 0.001 *** VS phylogenetic MANOVA approx-F9 = 3.05 < 0.001 *** forest shade PERMANOVA F9 = 5.38 0.001 *** UVS phylogenetic MANOVA approx-F9 = 3.07 < 0.001 *** against the sky PERMANOVA F9 = 5.38 0.001 *** VS phylogenetic MANOVA approx-F9 = 3.36 < 0.001 *** PERMANOVA F9 = 7.04 0.001 *** UVS phylogenetic MANOVA approx-F9 = 3.01 < 0.001 *** against a leaf PERMANOVA F9 = 7.07 0.001 *** VS phylogenetic MANOVA approx-F9 = 3.10 < 0.001 *** xyzL woodland shade PERMANOVA F9 = 5.33 0.001 *** UVS phylogenetic MANOVA approx-F9 = 3.12 < 0.001 *** against the sky PERMANOVA F9 = 5.34 0.002 ** VS phylogenetic MANOVA approx-F9 = 3.39 < 0.001 *** PERMANOVA F9 = 7.24 0.001 *** UVS phylogenetic MANOVA approx-F9 = 3.00 < 0.001 *** against a leaf PERMANOVA F9 = 7.24 0.001 *** VS phylogenetic MANOVA approx-F9 = 3.07 < 0.001 *** large gap PERMANOVA F9 = 5.37 0.001 *** UVS phylogenetic MANOVA approx-F9 = 3.14 < 0.001 *** against the sky PERMANOVA F9 = 5.37 0.001 *** VS phylogenetic MANOVA approx-F9 = 3.38 < 0.001 *** x, y and z are the mean coordinates in the tetrahedral colour space of transparent areas for each species and L is the mean luminance. -

Re-Emergence and Diversification of a Specialised Antennal Lobe Morphology in Ithomiine Butterflies
bioRxiv preprint doi: https://doi.org/10.1101/2020.10.13.336206; this version posted October 13, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Re-emergence and diversification of a specialised antennal lobe morphology 2 in ithomiine butterflies 3 4 Authors: 5 Billy J Morris1*, Antoine Couto1,2, Asli Aydin3, Stephen H Montgomery2*. 6 7 Affiliations: 1 8 Dept. of Zoology, University of Cambridge, Downing Street, Cambridge, CB2 3EJ 9 2 School of Biological Sciences, University of Bristol, 24 Tyndall Avenue, Bristol, BS8 1TQ 10 3 School of Medicine, Koc University, Rumelifeneri Yolu 34450 Sarıyer / Istanbul, Turkey 11 12 * corresponding authors: 13 BJM: [email protected] 14 SHM: [email protected] 15 16 Abstract 17 How an organism’s sensory system functions is central to how it navigates its environment and 18 meets the behavioural challenges associated with survival and reproduction. Comparing sensory 19 systems across species can reveal how facets of behaviour and ecology promote adaptive shifts 20 in the relative importance of certain environmental cues. The insect olfactory system is prominent 21 model for investigating how ecological factors impact sensory reception and processing. Notably 22 work in Lepidoptera led to the discovery of vastly expanded structures, termed a macroglomerular 23 complex (MGC), within the primary olfactory processing centre. These structures typically process 24 pheromonal cues and provide a classic example of how variation in size can influence the 25 functional processing of sensory cues. -

Rev Iss Web Mec 13773 25-22 5765..5784
Molecular Ecology (2016) 25, 5765–5784 doi: 10.1111/mec.13773 Into the Andes: multiple independent colonizations drive montane diversity in the Neotropical clearwing butterflies Godyridina NICOLAS CHAZOT,*† KEITH R. WILLMOTT,‡ FABIEN L. CONDAMINE,§¶ DONNA LISA DE-SILVA,* ANDREV.L.FREITAS,**GERARDOLAMAS,†† HELENE MORLON,‡‡ CARLOS E. GIRALDO,§§ CHRIS D. JIGGINS,¶¶ MATHIEU JORON,*** JAMES MALLET,††† SANDRA URIBE‡‡‡ and MARIANNE ELIAS* *Institut de Systematique, Evolution, Biodiversite, ISYEB – UMR 7205 – CNRS MNHN UPMC EPHE, Museum national d’Histoire naturelle, Sorbonne Universites, 57 rue Cuvier CP50, F-75005 Paris, France, †Department of Biology, University of Lund, 223 62 Lund, Sweden, ‡McGuire Center for Lepidoptera and Biodiversity, Florida Museum of Natural History, University of Florida, Gainesville, FL 32611, USA, §CNRS, UMR 5554 Institut des Sciences de l’Evolution (Universite de Montpellier), Place Eugene Bataillon, 34095 Montpellier, France, ¶Department of Biological Sciences, University of Alberta, T6G 2E9 Edmonton, AB, Canada, **Departamento de Zoologia and Museu de Zoologia, Instituto de Biologia, Universidade Estadual de Campinas, Campinas, S~ao Paulo, Brazil, ††Museo de Historia Natural, Universidad Nacional de San Marcos, Lima, Peru, ‡‡IBENS, Ecole Normale Superieure, UMR 8197 CNRS, Paris, France, §§Grupo de Investigacion de Sanidad Vegetal, Universidad Catolica de Oriente, Rionegro, Antioquia, Colombia, ¶¶Department of Zoology, University of Cambridge, Cambridge, UK, ***Centre d’Ecologie Fonctionnelle et Evolutive, CEFE, UMR 5175 CNRS – EPHE – Universite de Montpellier – Universite Paul Valery Montpellier, 34293 Montpellier 5, France, †††Department of Organismic and Evolutionary Biology, Harvard University, Cambridge, MA 02138, USA, ‡‡‡Universidad Nacional de Colombia, sede Medellın, Medellın, Colombia Abstract Understanding why species richness peaks along the Andes is a fundamental question in the study of Neotropical biodiversity.