Keratin Profiling by Single-Cell RNA-Sequencing Identifies Human
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Universidade Estadual De Campinas Instituto De Biologia
UNIVERSIDADE ESTADUAL DE CAMPINAS INSTITUTO DE BIOLOGIA VERÔNICA APARECIDA MONTEIRO SAIA CEREDA O PROTEOMA DO CORPO CALOSO DA ESQUIZOFRENIA THE PROTEOME OF THE CORPUS CALLOSUM IN SCHIZOPHRENIA CAMPINAS 2016 1 VERÔNICA APARECIDA MONTEIRO SAIA CEREDA O PROTEOMA DO CORPO CALOSO DA ESQUIZOFRENIA THE PROTEOME OF THE CORPUS CALLOSUM IN SCHIZOPHRENIA Dissertação apresentada ao Instituto de Biologia da Universidade Estadual de Campinas como parte dos requisitos exigidos para a obtenção do Título de Mestra em Biologia Funcional e Molecular na área de concentração de Bioquímica. Dissertation presented to the Institute of Biology of the University of Campinas in partial fulfillment of the requirements for the degree of Master in Functional and Molecular Biology, in the area of Biochemistry. ESTE ARQUIVO DIGITAL CORRESPONDE À VERSÃO FINAL DA DISSERTAÇÃO DEFENDIDA PELA ALUNA VERÔNICA APARECIDA MONTEIRO SAIA CEREDA E ORIENTADA PELO DANIEL MARTINS-DE-SOUZA. Orientador: Daniel Martins-de-Souza CAMPINAS 2016 2 Agência(s) de fomento e nº(s) de processo(s): CNPq, 151787/2F2014-0 Ficha catalográfica Universidade Estadual de Campinas Biblioteca do Instituto de Biologia Mara Janaina de Oliveira - CRB 8/6972 Saia-Cereda, Verônica Aparecida Monteiro, 1988- Sa21p O proteoma do corpo caloso da esquizofrenia / Verônica Aparecida Monteiro Saia Cereda. – Campinas, SP : [s.n.], 2016. Orientador: Daniel Martins de Souza. Dissertação (mestrado) – Universidade Estadual de Campinas, Instituto de Biologia. 1. Esquizofrenia. 2. Espectrometria de massas. 3. Corpo caloso. -

Propranolol-Mediated Attenuation of MMP-9 Excretion in Infants with Hemangiomas
Supplementary Online Content Thaivalappil S, Bauman N, Saieg A, Movius E, Brown KJ, Preciado D. Propranolol-mediated attenuation of MMP-9 excretion in infants with hemangiomas. JAMA Otolaryngol Head Neck Surg. doi:10.1001/jamaoto.2013.4773 eTable. List of All of the Proteins Identified by Proteomics This supplementary material has been provided by the authors to give readers additional information about their work. © 2013 American Medical Association. All rights reserved. Downloaded From: https://jamanetwork.com/ on 10/01/2021 eTable. List of All of the Proteins Identified by Proteomics Protein Name Prop 12 mo/4 Pred 12 mo/4 Δ Prop to Pred mo mo Myeloperoxidase OS=Homo sapiens GN=MPO 26.00 143.00 ‐117.00 Lactotransferrin OS=Homo sapiens GN=LTF 114.00 205.50 ‐91.50 Matrix metalloproteinase‐9 OS=Homo sapiens GN=MMP9 5.00 36.00 ‐31.00 Neutrophil elastase OS=Homo sapiens GN=ELANE 24.00 48.00 ‐24.00 Bleomycin hydrolase OS=Homo sapiens GN=BLMH 3.00 25.00 ‐22.00 CAP7_HUMAN Azurocidin OS=Homo sapiens GN=AZU1 PE=1 SV=3 4.00 26.00 ‐22.00 S10A8_HUMAN Protein S100‐A8 OS=Homo sapiens GN=S100A8 PE=1 14.67 30.50 ‐15.83 SV=1 IL1F9_HUMAN Interleukin‐1 family member 9 OS=Homo sapiens 1.00 15.00 ‐14.00 GN=IL1F9 PE=1 SV=1 MUC5B_HUMAN Mucin‐5B OS=Homo sapiens GN=MUC5B PE=1 SV=3 2.00 14.00 ‐12.00 MUC4_HUMAN Mucin‐4 OS=Homo sapiens GN=MUC4 PE=1 SV=3 1.00 12.00 ‐11.00 HRG_HUMAN Histidine‐rich glycoprotein OS=Homo sapiens GN=HRG 1.00 12.00 ‐11.00 PE=1 SV=1 TKT_HUMAN Transketolase OS=Homo sapiens GN=TKT PE=1 SV=3 17.00 28.00 ‐11.00 CATG_HUMAN Cathepsin G OS=Homo -

A Cell Line P53 Mutation Type UM
A Cell line p53 mutation Type UM-SCC 1 wt UM-SCC5 Exon 5, 157 GTC --> TTC Missense mutation by transversion (Valine --> Phenylalanine UM-SCC6 wt UM-SCC9 wt UM-SCC11A wt UM-SCC11B Exon 7, 242 TGC --> TCC Missense mutation by transversion (Cysteine --> Serine) UM-SCC22A Exon 6, 220 TAT --> TGT Missense mutation by transition (Tyrosine --> Cysteine) UM-SCC22B Exon 6, 220 TAT --> TGT Missense mutation by transition (Tyrosine --> Cysteine) UM-SCC38 Exon 5, 132 AAG --> AAT Missense mutation by transversion (Lysine --> Asparagine) UM-SCC46 Exon 8, 278 CCT --> CGT Missense mutation by transversion (Proline --> Alanine) B 1 Supplementary Methods Cell Lines and Cell Culture A panel of ten established HNSCC cell lines from the University of Michigan series (UM-SCC) was obtained from Dr. T. E. Carey at the University of Michigan, Ann Arbor, MI. The UM-SCC cell lines were derived from eight patients with SCC of the upper aerodigestive tract (supplemental Table 1). Patient age at tumor diagnosis ranged from 37 to 72 years. The cell lines selected were obtained from patients with stage I-IV tumors, distributed among oral, pharyngeal and laryngeal sites. All the patients had aggressive disease, with early recurrence and death within two years of therapy. Cell lines established from single isolates of a patient specimen are designated by a numeric designation, and where isolates from two time points or anatomical sites were obtained, the designation includes an alphabetical suffix (i.e., "A" or "B"). The cell lines were maintained in Eagle's minimal essential media supplemented with 10% fetal bovine serum and penicillin/streptomycin. -

Transcriptional Mechanisms of Resistance to Anti-PD-1 Therapy
Author Manuscript Published OnlineFirst on February 13, 2017; DOI: 10.1158/1078-0432.CCR-17-0270 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. Transcriptional mechanisms of resistance to anti-PD-1 therapy Maria L. Ascierto1, Alvin Makohon-Moore2, 11, Evan J. Lipson1, Janis M. Taube3,4, Tracee L. McMiller5, Alan E. Berger6, Jinshui Fan6, Genevieve J. Kaunitz3, Tricia R. Cottrell4, Zachary A. Kohutek7, Alexander Favorov8,10, Vladimir Makarov7,11, Nadeem Riaz7,11, Timothy A. Chan7,11, Leslie Cope8, Ralph H. Hruban4,9, Drew M. Pardoll1, Barry S. Taylor11,12,13, David B. Solit13, Christine A Iacobuzio-Donahue2,11, and Suzanne L. Topalian5 From the 1Departments of Oncology, 3Dermatology, 4Pathology, 5Surgery, 6The Lowe Family Genomics Core, 8Oncology Bioinformatics Core, and the 9 Sol Goldman Pancreatic Cancer Research Center, Johns Hopkins University School of Medicine and Sidney Kimmel Comprehensive Cancer Center, Baltimore, MD 21287; the 10Laboratory of System Biology and Computational Genetics, Vavilov Institute of General Genetics, Russian Academy of Sciences, 119991, Moscow, Russia; and 2Pathology, 7Radiation Oncology, 11Human Oncology and Pathogenesis Program, 12Department of Epidemiology and Biostatistics, and the 13Center for Molecular Oncology, Memorial Sloan Kettering Cancer Center, New York NY 10065. MLA, AM-M, EJL, and JMT contributed equally to this work Running title: Transcriptional mechanisms of resistance to anti-PD-1 Key Words: melanoma, cancer genetics, immunotherapy, anti-PD-1 Financial Support: This study was supported by the Melanoma Research Alliance (to SLT and CI-D), the Bloomberg~Kimmel Institute for Cancer Immunotherapy (to JMT, DMP, and SLT), the Barney Family Foundation (to SLT), Moving for Melanoma of Delaware (to SLT), the 1 Downloaded from clincancerres.aacrjournals.org on October 2, 2021. -

Steroid-Dependent Regulation of the Oviduct: a Cross-Species Transcriptomal Analysis
University of Kentucky UKnowledge Theses and Dissertations--Animal and Food Sciences Animal and Food Sciences 2015 Steroid-dependent regulation of the oviduct: A cross-species transcriptomal analysis Katheryn L. Cerny University of Kentucky, [email protected] Right click to open a feedback form in a new tab to let us know how this document benefits ou.y Recommended Citation Cerny, Katheryn L., "Steroid-dependent regulation of the oviduct: A cross-species transcriptomal analysis" (2015). Theses and Dissertations--Animal and Food Sciences. 49. https://uknowledge.uky.edu/animalsci_etds/49 This Doctoral Dissertation is brought to you for free and open access by the Animal and Food Sciences at UKnowledge. It has been accepted for inclusion in Theses and Dissertations--Animal and Food Sciences by an authorized administrator of UKnowledge. For more information, please contact [email protected]. STUDENT AGREEMENT: I represent that my thesis or dissertation and abstract are my original work. Proper attribution has been given to all outside sources. I understand that I am solely responsible for obtaining any needed copyright permissions. I have obtained needed written permission statement(s) from the owner(s) of each third-party copyrighted matter to be included in my work, allowing electronic distribution (if such use is not permitted by the fair use doctrine) which will be submitted to UKnowledge as Additional File. I hereby grant to The University of Kentucky and its agents the irrevocable, non-exclusive, and royalty-free license to archive and make accessible my work in whole or in part in all forms of media, now or hereafter known. -

Plakoglobin Is Required for Effective Intermediate Filament Anchorage to Desmosomes Devrim Acehan1, Christopher Petzold1, Iwona Gumper2, David D
ORIGINAL ARTICLE Plakoglobin Is Required for Effective Intermediate Filament Anchorage to Desmosomes Devrim Acehan1, Christopher Petzold1, Iwona Gumper2, David D. Sabatini2, Eliane J. Mu¨ller3, Pamela Cowin2,4 and David L. Stokes1,2,5 Desmosomes are adhesive junctions that provide mechanical coupling between cells. Plakoglobin (PG) is a major component of the intracellular plaque that serves to connect transmembrane elements to the cytoskeleton. We have used electron tomography and immunolabeling to investigate the consequences of PG knockout on the molecular architecture of the intracellular plaque in cultured keratinocytes. Although knockout keratinocytes form substantial numbers of desmosome-like junctions and have a relatively normal intercellular distribution of desmosomal cadherins, their cytoplasmic plaques are sparse and anchoring of intermediate filaments is defective. In the knockout, b-catenin appears to substitute for PG in the clustering of cadherins, but is unable to recruit normal levels of plakophilin-1 and desmoplakin to the plaque. By comparing tomograms of wild type and knockout desmosomes, we have assigned particular densities to desmoplakin and described their interaction with intermediate filaments. Desmoplakin molecules are more extended in wild type than knockout desmosomes, as if intermediate filament connections produced tension within the plaque. On the basis of our observations, we propose a particular assembly sequence, beginning with cadherin clustering within the plasma membrane, followed by recruitment of plakophilin and desmoplakin to the plaque, and ending with anchoring of intermediate filaments, which represents the key to adhesive strength. Journal of Investigative Dermatology (2008) 128, 2665–2675; doi:10.1038/jid.2008.141; published online 22 May 2008 INTRODUCTION dense plaque that is further from the membrane and that Desmosomes are large macromolecular complexes that mediates the binding of intermediate filaments. -

The Act of Controlling Adult Stem Cell Dynamics: Insights from Animal Models
biomolecules Review The Act of Controlling Adult Stem Cell Dynamics: Insights from Animal Models Meera Krishnan 1, Sahil Kumar 1, Luis Johnson Kangale 2,3 , Eric Ghigo 3,4 and Prasad Abnave 1,* 1 Regional Centre for Biotechnology, NCR Biotech Science Cluster, 3rd Milestone, Gurgaon-Faridabad Ex-pressway, Faridabad 121001, India; [email protected] (M.K.); [email protected] (S.K.) 2 IRD, AP-HM, SSA, VITROME, Aix-Marseille University, 13385 Marseille, France; [email protected] 3 Institut Hospitalo Universitaire Méditerranée Infection, 13385 Marseille, France; [email protected] 4 TechnoJouvence, 13385 Marseille, France * Correspondence: [email protected] Abstract: Adult stem cells (ASCs) are the undifferentiated cells that possess self-renewal and differ- entiation abilities. They are present in all major organ systems of the body and are uniquely reserved there during development for tissue maintenance during homeostasis, injury, and infection. They do so by promptly modulating the dynamics of proliferation, differentiation, survival, and migration. Any imbalance in these processes may result in regeneration failure or developing cancer. Hence, the dynamics of these various behaviors of ASCs need to always be precisely controlled. Several genetic and epigenetic factors have been demonstrated to be involved in tightly regulating the proliferation, differentiation, and self-renewal of ASCs. Understanding these mechanisms is of great importance, given the role of stem cells in regenerative medicine. Investigations on various animal models have played a significant part in enriching our knowledge and giving In Vivo in-sight into such ASCs regulatory mechanisms. In this review, we have discussed the recent In Vivo studies demonstrating the role of various genetic factors in regulating dynamics of different ASCs viz. -

Molecular Pathways Associated with the Nutritional Programming of Plant-Based Diet Acceptance in Rainbow Trout Following an Early Feeding Exposure Mukundh N
Molecular pathways associated with the nutritional programming of plant-based diet acceptance in rainbow trout following an early feeding exposure Mukundh N. Balasubramanian, Stéphane Panserat, Mathilde Dupont-Nivet, Edwige Quillet, Jérôme Montfort, Aurélie Le Cam, Françoise Médale, Sadasivam J. Kaushik, Inge Geurden To cite this version: Mukundh N. Balasubramanian, Stéphane Panserat, Mathilde Dupont-Nivet, Edwige Quillet, Jérôme Montfort, et al.. Molecular pathways associated with the nutritional programming of plant-based diet acceptance in rainbow trout following an early feeding exposure. BMC Genomics, BioMed Central, 2016, 17 (1), pp.1-20. 10.1186/s12864-016-2804-1. hal-01346912 HAL Id: hal-01346912 https://hal.archives-ouvertes.fr/hal-01346912 Submitted on 19 Jul 2016 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Balasubramanian et al. BMC Genomics (2016) 17:449 DOI 10.1186/s12864-016-2804-1 RESEARCH ARTICLE Open Access Molecular pathways associated with the nutritional programming of plant-based diet acceptance in rainbow trout following an early feeding exposure Mukundh N. Balasubramanian1, Stephane Panserat1, Mathilde Dupont-Nivet2, Edwige Quillet2, Jerome Montfort3, Aurelie Le Cam3, Francoise Medale1, Sadasivam J. Kaushik1 and Inge Geurden1* Abstract Background: The achievement of sustainable feeding practices in aquaculture by reducing the reliance on wild-captured fish, via replacement of fish-based feed with plant-based feed, is impeded by the poor growth response seen in fish fed high levels of plant ingredients. -

Analysis of Mouse Keratin 6A Regulatory Sequences in Transgenic Mice Reveals Constitutive, Tissue-Speci®C Expression by a Keratin 6A Minigene
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Analysis of Mouse Keratin 6a Regulatory Sequences in Transgenic Mice Reveals Constitutive, Tissue-Speci®c Expression by a Keratin 6a Minigene Donna Mahony, Seetha Karunaratne, Graham Cam,* and Joseph A. Rothnagel Department of Biochemistry and the Institute for Molecular Bioscience, University of Queensland, Brisbane, Queensland, and *Division of Animal Production, CSIRO, Blacktown, New South Wales, Australia The analysis of keratin 6 expression is complicated atin 6 expressing tissues, including the hair follicle, by the presence of multiple isoforms that are tongue, footpad, and nail bed, showing that both expressed constitutively in a number of internal stra- transgenes retained keratinocyte-speci®c expression. ti®ed epithelia, in palmoplantar epidermis, and in Quantitative analysis of b-galactosidase activity veri- the companion cell layer of the hair follicle. In addi- ®ed that both the 1.3 and 0.12 kb keratin 6a promo- tion, keratin 6 expression is inducible in interfollicu- ter constructs produced similar levels of the reporter. lar epidermis and the outer root sheath of the Notably, bothconstructs were constitutively follicle, in response to wounding stimuli, phorbol expressed in the outer root sheath and interfollicular esters, or retinoic acid. In order to establishthecriti- epidermis in the absence of any activating stimulus, cal regions involved in the regulation of keratin 6a suggesting that they lack the regulatory elements (the dominant isoform in mice), we generated trans- that normally silence transcription in these cells. This genic mice withtwo different-sized mouse keratin 6a study has revealed that a keratin 6a minigene con- constructs containing either 1.3 kb or 0.12 kb of 5¢ tains critical cis elements that mediate tissue-speci®c ¯anking sequence linked to the lacZ reporter gene. -
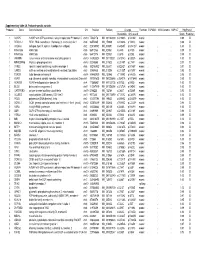
Supplementary Table 2A. Proband-Specific Variants Proband
Supplementary table 2A. Proband-specific variants Proband Gene Gene full name Chr Position RefSeqChange Function ESP6500 1000 Genome SNP137 PolyPhen2 Nucleotide Amino acid Score Prediction 1 AGAP5 ArfGAP with GTPase domain, ankyrin repeat and PH domain 5 chr10 75435178 NM_001144000 c.G1240A p.V414M exonic . 0.99 D 1 REXO1L1 REX1, RNA exonuclease 1 homolog (S. cerevisiae)-like 1 chr8 86574465 NM_172239 c.A1262G p.Y421C exonic . 0.98 D 1 COL4A3 collagen, type IV, alpha 3 (Goodpasture antigen) chr2 228169782 NM_000091 c.G4235T p.G1412V exonic . 1.00 D 1 KIAA1586 KIAA1586 chr6 56912133 NM_020931 c.C44G p.A15G exonic . 0.99 D 1 KIAA1586 KIAA1586 chr6 56912176 NM_020931 c.C87G p.D29E exonic . 0.99 D 1 JAKMIP3 Janus kinase and microtubule interacting protein 3 chr10 133955524 NM_001105521 c.G1574C p.G525A exonic . 1.00 D 1 MPHOSPH8 M-phase phosphoprotein 8 chr13 20240685 NM_017520 c.C2140T p.L714F exonic . 0.97 D 1 SYNE1 spectrin repeat containing, nuclear envelope 1 chr6 152783902 NM_033071 c.A2242T p.N748Y exonic . 0.97 D 1 CAND2 cullin-associated and neddylation-dissociated 2 (putative) chr3 12858835 NM_012298 c.C2125T p.H709Y exonic . 0.95 D 1 TDRD9 tudor domain containing 9 chr14 104460909 NM_153046 c.T1289G p.V430G exonic . 0.96 D 2 ASPM asp (abnormal spindle) homolog, microcephaly associated (Drosophila)chr1 197091625 NM_001206846 c.G3491A p.R1164H exonic . 0.99 D 2 ADAM29 ADAM metallopeptidase domain 29 chr4 175896951 NM_001130705 c.A275G p.D92G exonic . 1.00 D 2 BCO2 beta-carotene oxygenase 2 chr11 112087019 NM_001256398 c.G1373A p.G458E exonic . -

Structural and Biochemical Changes Underlying a Keratoderma-Like Phenotype in Mice Lacking Suprabasal AP1 Transcription Factor Function
Citation: Cell Death and Disease (2015) 6, e1647; doi:10.1038/cddis.2015.21 OPEN & 2015 Macmillan Publishers Limited All rights reserved 2041-4889/15 www.nature.com/cddis Structural and biochemical changes underlying a keratoderma-like phenotype in mice lacking suprabasal AP1 transcription factor function EA Rorke*,1, G Adhikary2, CA Young2, RH Rice3, PM Elias4, D Crumrine4, J Meyer4, M Blumenberg5 and RL Eckert2,6,7,8 Epidermal keratinocyte differentiation on the body surface is a carefully choreographed process that leads to assembly of a barrier that is essential for life. Perturbation of keratinocyte differentiation leads to disease. Activator protein 1 (AP1) transcription factors are key controllers of this process. We have shown that inhibiting AP1 transcription factor activity in the suprabasal murine epidermis, by expression of dominant-negative c-jun (TAM67), produces a phenotype type that resembles human keratoderma. However, little is understood regarding the structural and molecular changes that drive this phenotype. In the present study we show that TAM67-positive epidermis displays altered cornified envelope, filaggrin-type keratohyalin granule, keratin filament, desmosome formation and lamellar body secretion leading to reduced barrier integrity. To understand the molecular changes underlying this process, we performed proteomic and RNA array analysis. Proteomic study of the corneocyte cross-linked proteome reveals a reduction in incorporation of cutaneous keratins, filaggrin, filaggrin2, late cornified envelope precursor proteins, hair keratins and hair keratin-associated proteins. This is coupled with increased incorporation of desmosome linker, small proline-rich, S100, transglutaminase and inflammation-associated proteins. Incorporation of most cutaneous keratins (Krt1, Krt5 and Krt10) is reduced, but incorporation of hyperproliferation-associated epidermal keratins (Krt6a, Krt6b and Krt16) is increased. -

CDH12 Cadherin 12, Type 2 N-Cadherin 2 RPL5 Ribosomal
5 6 6 5 . 4 2 1 1 1 2 4 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 2 A A A A A A A A A A A A A A A A A A A A C C C C C C C C C C C C C C C C C C C C R R R R R R R R R R R R R R R R R R R R B , B B B B B B B B B B B B B B B B B B B , 9 , , , , 4 , , 3 0 , , , , , , , , 6 2 , , 5 , 0 8 6 4 , 7 5 7 0 2 8 9 1 3 3 3 1 1 7 5 0 4 1 4 0 7 1 0 2 0 6 7 8 0 2 5 7 8 0 3 8 5 4 9 0 1 0 8 8 3 5 6 7 4 7 9 5 2 1 1 8 2 2 1 7 9 6 2 1 7 1 1 0 4 5 3 5 8 9 1 0 0 4 2 5 0 8 1 4 1 6 9 0 0 6 3 6 9 1 0 9 0 3 8 1 3 5 6 3 6 0 4 2 6 1 0 1 2 1 9 9 7 9 5 7 1 5 8 9 8 8 2 1 9 9 1 1 1 9 6 9 8 9 7 8 4 5 8 8 6 4 8 1 1 2 8 6 2 7 9 8 3 5 4 3 2 1 7 9 5 3 1 3 2 1 2 9 5 1 1 1 1 1 1 5 9 5 3 2 6 3 4 1 3 1 1 4 1 4 1 7 1 3 4 3 2 7 6 4 2 7 2 1 2 1 5 1 6 3 5 6 1 3 6 4 7 1 6 5 1 1 4 1 6 1 7 6 4 7 e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m