The Complete Mitogenome of €Lysmata Vittata
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
W7192e19.Pdf
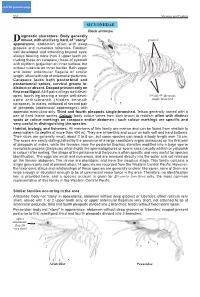
click for previous page 952 Shrimps and Prawns Sicyoniidae SICYONIIDAE Rock shrimps iagnostic characters: Body generally Drobust, with shell very hard, of “stony” grooves appearance; abdomen often with deep grooves and numerous tubercles. Rostrum well developed and extending beyond eyes, always bearing more than 3 upper teeth (in- cluding those on carapace); base of eyestalk with styliform projection on inner surface, but without tubercle on inner border. Both upper and lower antennular flagella of similar length, attached to tip of antennular peduncle. 1 Carapace lacks both postorbital and postantennal spines, cervical groove in- distinct or absent. Exopod present only on first maxilliped. All 5 pairs of legs well devel- 2 oped, fourth leg bearing a single well-devel- 3rd and 4th pleopods 4 single-branched oped arthrobranch (hidden beneath 3 carapace). In males, endopod of second pair 5 of pleopods (abdominal appendages) with appendix masculina only. Third and fourth pleopods single-branched. Telson generally armed with a pair of fixed lateral spines. Colour: body colour varies from dark brown to reddish; often with distinct spots or colour markings on carapace and/or abdomen - such colour markings are specific and very useful in distinguishing the species. Habitat, biology, and fisheries: All members of this family are marine and can be found from shallow to deep waters (to depths of more than 400 m). They are all benthic and occur on both soft and hard bottoms. Their sizes are generally small, about 2 to 8 cm, but some species can reach a body length over 15 cm. The sexes are easily distinguished by the presence of a large copulatory organ (petasma) on the first pair of pleopods of males, while the females have the posterior thoracic sternites modified into a large sperm receptacle process (thelycum) which holds the spermatophores or sperm sacs (usually whitish or yellowish in colour) after mating. -

The Isolation, Genetic Characterisation And
The isolation, genetic characterisation and biological activity of a South African Phthorimaea operculella granulovirus (PhopGV-SA) for the control of the Potato Tuber Moth, Phthorimaea operculella (Zeller) A thesis submitted in fulfilment of the requirements for the degree of MASTER OF SCIENCE At RHODES UNIVERSITY By MICHAEL DAVID JUKES February 2015 i Abstract The potato tuber moth, Phthorimaea operculella (Zeller), is a major pest of potato crops worldwide causing significant damage to both field and stored tubers. The current control method in South Africa involves chemical insecticides, however, there is growing concern on the health and environmental risks of their use. The development of novel biopesticide based control methods may offer a potential solution for the future of insecticides. In this study a baculovirus was successfully isolated from a laboratory population of P. operculella. Transmission electron micrographs revealed granulovirus-like particles. DNA was extracted from recovered occlusion bodies and used for the PCR amplification of the lef-8, lef- 9, granulin and egt genes. Sequence data was obtained and submitted to BLAST identifying the virus as a South African isolate of Phthorimaea operculella granulovirus (PhopGV-SA). Phylogenetic analysis of the lef-8, lef-9 and granulin amino acid sequences grouped the South African isolate with PhopGV-1346. Comparison of egt sequence data identified PhopGV-SA as a type II egt gene. A phylogenetic analysis of egt amino acid sequences grouped all type II genes, including PhopGV-SA, into a separate clade from types I, III, IV and V. These findings suggest that type II may represent the prototype structure for this gene with the evolution of types I, III and IV a result of large internal deletion events and subsequent divergence. -

Contribution to the Knowledge of the Fauna of Bombyces, Sphinges And
driemaandelijks tijdschrift van de VLAAMSE VERENIGING VOOR ENTOMOLOGIE Afgiftekantoor 2170 Merksem 1 ISSN 0771-5277 Periode: oktober – november – december 2002 Erkenningsnr. P209674 Redactie: Dr. J–P. Borie (Compiègne, France), Dr. L. De Bruyn (Antwerpen), T. C. Garrevoet (Antwerpen), B. Goater (Chandlers Ford, England), Dr. K. Maes (Gent), Dr. K. Martens (Brussel), H. van Oorschot (Amsterdam), D. van der Poorten (Antwerpen), W. O. De Prins (Antwerpen). Redactie-adres: W. O. De Prins, Nieuwe Donk 50, B-2100 Antwerpen (Belgium). e-mail: [email protected]. Jaargang 30, nummer 4 1 december 2002 Contribution to the knowledge of the fauna of Bombyces, Sphinges and Noctuidae of the Southern Ural Mountains, with description of a new Dichagyris (Lepidoptera: Lasiocampidae, Endromidae, Saturniidae, Sphingidae, Notodontidae, Noctuidae, Pantheidae, Lymantriidae, Nolidae, Arctiidae) Kari Nupponen & Michael Fibiger [In co-operation with Vladimir Olschwang, Timo Nupponen, Jari Junnilainen, Matti Ahola and Jari- Pekka Kaitila] Abstract. The list, comprising 624 species in the families Lasiocampidae, Endromidae, Saturniidae, Sphingidae, Notodontidae, Noctuidae, Pantheidae, Lymantriidae, Nolidae and Arctiidae from the Southern Ural Mountains is presented. The material was collected during 1996–2001 in 10 different expeditions. Dichagyris lux Fibiger & K. Nupponen sp. n. is described. 17 species are reported for the first time from Europe: Clostera albosigma (Fitch, 1855), Xylomoia retinax Mikkola, 1998, Ecbolemia misella (Püngeler, 1907), Pseudohadena stenoptera Boursin, 1970, Hadula nupponenorum Hacker & Fibiger, 2002, Saragossa uralica Hacker & Fibiger, 2002, Conisania arida (Lederer, 1855), Polia malchani (Draudt, 1934), Polia vespertilio (Draudt, 1934), Polia altaica (Lederer, 1853), Mythimna opaca (Staudinger, 1899), Chersotis stridula (Hampson, 1903), Xestia wockei (Möschler, 1862), Euxoa dsheiron Brandt, 1938, Agrotis murinoides Poole, 1989, Agrotis sp. -

SPG2: Biodiversity Conservation (July 2006) 1 1.0 an OVERVIEW
Kent and Medway Structure Plan 2006 mapping out the future Supplementary Planning Guidance SPG2 Biodiversity Conservation July 2006 Strategy and Planning Division/ Environment and Waste Division Environment and Regeneration Directorate Kent County Council Tel: 01622 221609 Email: [email protected] Kent and Medway Structure Plan 2006 Supplementary Planning Guidance (SPG2): Biodiversity Conservation Preface i. The purpose of Supplementary Planning Guidance (SPG) is to supplement the policies and proposals of development plans. It elaborates policies so that they can be better understood and effectively applied. SPG should be clearly cross-referenced to the relevant plan policy or policies which it supplements and should be the subject of consultation during its preparation. In these circumstances SPG may be taken into account as a material consideration in planning decisions. ii. A number of elements of SPG have been produced to supplement certain policies in the Kent and Medway Structure Plan. This SPG supplements the following policies: • Policy EN6: International and National Wildlife Designations • Policy EN7: County and Local Wildlife Designations • Policy EN8: Protecting, Conserving and Enhancing Biodiversity • Policy EN9: Trees, Woodland and Hedgerows iii. This SPG has been prepared by Kent County Council working in partnership with a range of stakeholders drawn from Kent local authorities and other relevant agencies. iv. A draft of this SPG was subject to public consultation alongside public consultation on the deposit draft of the Kent and Medway Structure Plan in late 2003. It has been subsequently revised and updated prior to its adoption. A separate report provides a statement of the consultation undertaken, the representations received and the response to these representations. -

Download-The-Data (Accessed on 12 July 2021))
diversity Article Integrative Taxonomy of New Zealand Stenopodidea (Crustacea: Decapoda) with New Species and Records for the Region Kareen E. Schnabel 1,* , Qi Kou 2,3 and Peng Xu 4 1 Coasts and Oceans Centre, National Institute of Water & Atmospheric Research, Private Bag 14901 Kilbirnie, Wellington 6241, New Zealand 2 Institute of Oceanology, Chinese Academy of Sciences, Qingdao 266071, China; [email protected] 3 College of Marine Science, University of Chinese Academy of Sciences, Beijing 100049, China 4 Key Laboratory of Marine Ecosystem Dynamics, Second Institute of Oceanography, Ministry of Natural Resources, Hangzhou 310012, China; [email protected] * Correspondence: [email protected]; Tel.: +64-4-386-0862 Abstract: The New Zealand fauna of the crustacean infraorder Stenopodidea, the coral and sponge shrimps, is reviewed using both classical taxonomic and molecular tools. In addition to the three species so far recorded in the region, we report Spongicola goyi for the first time, and formally describe three new species of Spongicolidae. Following the morphological review and DNA sequencing of type specimens, we propose the synonymy of Spongiocaris yaldwyni with S. neocaledonensis and review a proposed broad Indo-West Pacific distribution range of Spongicoloides novaezelandiae. New records for the latter at nearly 54◦ South on the Macquarie Ridge provide the southernmost record for stenopodidean shrimp known to date. Citation: Schnabel, K.E.; Kou, Q.; Xu, Keywords: sponge shrimp; coral cleaner shrimp; taxonomy; cytochrome oxidase 1; 16S ribosomal P. Integrative Taxonomy of New RNA; association; southwest Pacific Ocean Zealand Stenopodidea (Crustacea: Decapoda) with New Species and Records for the Region. -

Effects of Exogenous Methyl Jasmonate-Induced Resistance In
Effects of exogenous methyl jasmonate-induced resistance in Populus × euramericana ‘Nanlin895’ on the performance and metabolic enzyme activities of Clostera anachoreta Gu Tianzi, Zhang Congcong, Chen Changyu, Li hui, Huang kairu, Tian Shuo, Zhao Xudong & Hao Dejun Arthropod-Plant Interactions An international journal devoted to studies on interactions of insects, mites, and other arthropods with plants ISSN 1872-8855 Volume 12 Number 2 Arthropod-Plant Interactions (2018) 12:247-255 DOI 10.1007/s11829-017-9564-y 1 23 Your article is protected by copyright and all rights are held exclusively by Springer Science+Business Media B.V.. This e-offprint is for personal use only and shall not be self- archived in electronic repositories. If you wish to self-archive your article, please use the accepted manuscript version for posting on your own website. You may further deposit the accepted manuscript version in any repository, provided it is only made publicly available 12 months after official publication or later and provided acknowledgement is given to the original source of publication and a link is inserted to the published article on Springer's website. The link must be accompanied by the following text: "The final publication is available at link.springer.com”. 1 23 Author's personal copy Arthropod-Plant Interactions (2018) 12:247–255 https://doi.org/10.1007/s11829-017-9564-y ORIGINAL PAPER Effects of exogenous methyl jasmonate-induced resistance in Populus 3 euramericana ‘Nanlin895’ on the performance and metabolic enzyme activities of Clostera anachoreta 1,2 1,2 1,2 1,2 1,2 Gu Tianzi • Zhang Congcong • Chen Changyu • Li hui • Huang kairu • 1,2 1,2 1,2 Tian Shuo • Zhao Xudong • Hao Dejun Received: 8 October 2016 / Accepted: 7 September 2017 / Published online: 21 September 2017 Ó Springer Science+Business Media B.V. -

Midgut Transcriptome Analysis of Clostera Anachoreta Treated with Cry1ac Toxin
bioRxiv preprint doi: https://doi.org/10.1101/568337; this version posted March 5, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Midgut Transcriptome Analysis of Clostera anachoreta Treated with Cry1Ac Toxin Liu Jie, Wang Liucheng, Zhou Guona, Liu Junxia, Gao Baojia Forestry College, Agricultural University of Hebei, 071000, Baoding, PR China. Correspondence and requests for materials should be addressed to (email: [email protected]) Abstract: Clostera anachoreta is one of the important Lepidoptera insect pests in forestry, especially in poplars woods in China, Europe, Japan and India, et al, and also the target insect of Cry1Ac toxin and Bt plants. In this study, by using the different dosages of Btcry1Ac toxin to feed larvae and analyzing the transcriptome data, we found six genes, HSC70, GNB2L/RACK1, PNLIP, BI1-like, arylphorin type 2 and PKM, might be associated with Cry1Ac toxin. And PI3K-Akt pathway, which was highly enriched in DEGs and linked to several crucial pathways, including the B cell receptor signaling pathway, toll-like receptor pathway, and MAPK signaling pathway, might be involved in the recovery stage when response to sub-lethal Cry1Ac toxin. This is the first study conducted to specifically investigate C. anachoreta response to Cry toxin stress using large-scale sequencing technologies, and the results highlighted some important genes and pathway that could be involved in Btcry1Ac resistance development or could serve as targets for biologically-based control mechanisms of this insect pest. -

Annotated Checklist of New Zealand Decapoda (Arthropoda: Crustacea)
Tuhinga 22: 171–272 Copyright © Museum of New Zealand Te Papa Tongarewa (2011) Annotated checklist of New Zealand Decapoda (Arthropoda: Crustacea) John C. Yaldwyn† and W. Richard Webber* † Research Associate, Museum of New Zealand Te Papa Tongarewa. Deceased October 2005 * Museum of New Zealand Te Papa Tongarewa, PO Box 467, Wellington, New Zealand ([email protected]) (Manuscript completed for publication by second author) ABSTRACT: A checklist of the Recent Decapoda (shrimps, prawns, lobsters, crayfish and crabs) of the New Zealand region is given. It includes 488 named species in 90 families, with 153 (31%) of the species considered endemic. References to New Zealand records and other significant references are given for all species previously recorded from New Zealand. The location of New Zealand material is given for a number of species first recorded in the New Zealand Inventory of Biodiversity but with no further data. Information on geographical distribution, habitat range and, in some cases, depth range and colour are given for each species. KEYWORDS: Decapoda, New Zealand, checklist, annotated checklist, shrimp, prawn, lobster, crab. Contents Introduction Methods Checklist of New Zealand Decapoda Suborder DENDROBRANCHIATA Bate, 1888 ..................................... 178 Superfamily PENAEOIDEA Rafinesque, 1815.............................. 178 Family ARISTEIDAE Wood-Mason & Alcock, 1891..................... 178 Family BENTHESICYMIDAE Wood-Mason & Alcock, 1891 .......... 180 Family PENAEIDAE Rafinesque, 1815 .................................. -

World Bank Document
Public Disclosure Authorized Desertification Control and Ecological Conservation Project in Ningxia, China Pest Management Plan Public Disclosure Authorized Forestry Bureau of the Ningxia Hui Autonomous Region Public Disclosure Authorized October 10, 2011 Public Disclosure Authorized Contents 2.1 Occurrence of Pests and Diseases ................................................................3 2.2 Current Pest Control Methods in Ningxia ..................................................7 3.1 Pesticide regulatory framework in Ningxia ................................................9 3.2 Administration of pests/diseases control ...................................................10 3.3 Organizational Responsibilities for Quality and Safety .......................... 11 4.1 The basic principles .....................................................................................12 4.2 Improving pesticide use for forest pest control ........................................12 4.3 Management of pesticides distribution and use. ....................................13 4.4 Measures of pesticides use ........................................................................14 5.1 Purpose of an Integrated Pest Management (IPM) System ....................16 5.2 Main Forest Pest Control Methods and Pesticide Types Proposed ........17 5.2.1 Control methods .......................................................................................17 5.2.2 Pesticide types ...........................................................................................18 -

Crustacea: Decapoda) Can Penetrate the Abyss: a New Species of Lebbeus from the Sea of Okhotsk, Representing the Deepest Record of the Family
European Journal of Taxonomy 604: 1–35 ISSN 2118-9773 https://doi.org/10.5852/ejt.2020.604 www.europeanjournaloftaxonomy.eu 2020 · Marin I. This work is licensed under a Creative Commons Attribution Licence (CC BY 4.0). Research article urn:lsid:zoobank.org:pub:7F2F71AA-4282-477C-9D6A-4C5FB417259D Thoridae (Crustacea: Decapoda) can penetrate the Abyss: a new species of Lebbeus from the Sea of Okhotsk, representing the deepest record of the family Ivan MARIN A.N. Severtzov Institute of Ecology and Evolution, Russian Academy of Sciences, Moscow, Russia. Email: [email protected], [email protected] urn:lsid:zoobank.org:author:B26ADAA5-5DBE-42B3-9784-3BC362540034 Abstract. Lebbeus sokhobio sp. nov. is described from abyssal depths (3303−3366 m) in the Kuril Basin of the Sea of Okhotsk. The related congeners are deep-water dwellers with a very distant distribution and very similar morphology. The new species is separated by minor morphological features, such as the armature of the rostrum and telson, meral spinulation of ambulatory pereiopods and the shape of the pleonal pleurae. This species is the deepest dwelling representative of the genus Lebbeus and the family Thoridae. A list of records of caridean shrimps recorded from abyssal depths below 3000 m is given. Keywords. Diversity, Caridea, barcoding, SokhoBio 2015, NW Pacifi c. Marin I. 2020. Thoridae (Crustacea: Decapoda) can penetrate the Abyss: a new species of Lebbeus from the Sea of Okhotsk, representing the deepest record of the family. European Journal of Taxonomy 604: 1–35. https://doi.org/10.5852/ejt.2020.604 Introduction The fauna of benthic caridean shrimps (Crustacea: Decapoda: Caridea) living at depths of more than 3000 m is poorly known due to the technical diffi culties of sampling. -

A Morphological and Molecular Study of Diversity and Connectivity Among Anchialine Shrimp
Florida International University FIU Digital Commons FIU Electronic Theses and Dissertations University Graduate School 11-10-2020 Connections in the Underworld: A Morphological and Molecular Study of Diversity and Connectivity among Anchialine Shrimp. Robert Eugene Ditter Florida International University, [email protected] Follow this and additional works at: https://digitalcommons.fiu.edu/etd Part of the Biodiversity Commons, Bioinformatics Commons, Biology Commons, Ecology and Evolutionary Biology Commons, Genetics and Genomics Commons, Marine Biology Commons, Natural Resources and Conservation Commons, Oceanography Commons, Other Environmental Sciences Commons, Speleology Commons, and the Zoology Commons Recommended Citation Ditter, Robert Eugene, "Connections in the Underworld: A Morphological and Molecular Study of Diversity and Connectivity among Anchialine Shrimp." (2020). FIU Electronic Theses and Dissertations. 4561. https://digitalcommons.fiu.edu/etd/4561 This work is brought to you for free and open access by the University Graduate School at FIU Digital Commons. It has been accepted for inclusion in FIU Electronic Theses and Dissertations by an authorized administrator of FIU Digital Commons. For more information, please contact [email protected]. FLORIDA INTERNATIONAL UNIVERSITY Miami, Florida CONNECTIONS IN THE UNDERWORLD: A MORPHOLOGICAL AND MOLECULAR STUDY OF DIVERSITY AND CONNECTIVITY AMONG ANCHIALINE SHRIMP A dissertation submitted in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY in BIOLOGY by Robert E. Ditter 2020 To: Dean Michael R. Heithaus College of Arts, Sciences and Education This dissertation, written by Robert E. Ditter, and entitled Connections in the Underworld: A Morphological and Molecular Study of Diversity and Connectivity among Anchialine Shrimp, having been approved in respect to style and intellectual content, is referred to you for judgment. -

British Lepidoptera (/)
British Lepidoptera (/) Home (/) Anatomy (/anatomy.html) FAMILIES 1 (/families-1.html) GELECHIOIDEA (/gelechioidea.html) FAMILIES 3 (/families-3.html) FAMILIES 4 (/families-4.html) NOCTUOIDEA (/noctuoidea.html) BLOG (/blog.html) Family: NOTODONTIDAE (6SF 16G+1EX 27S+1EX+1CI) Suborder:Glossata Infraorder:Heteroneura Superfamily:Noctuoidea References: Waring & Townsend, Wikipedia Long-winged, heavy-bodied; wings held in tectiform position at rest; antennae bipectinate in male, with shorter pectinations or ciliate in female; adults do not feed and proboscis may be developed, rudimentary or absent; tympanal organ on metathorax; tibial spurs serrate. Subfamily: Thaumatopoeinae are the 'Processionary' moths - reflecting the fact that the larvae leave their silken webs in procession at night to feed. They have previously been treated as a separate family (Thaumatopoidae) or within the family: Noctuidae Subfamily: Crerurinae include the Puss Moth - reflecting its hairiness, and the 'Kittens' - which resemble small Puss Moths Subfamily: Notodontinae are the 'Prominents' - referring to a projecting scale-tooth from the forewing dorsum (notodonta = "tooth-back") Hindtibia with only one pair of spurs in subfamilies Cerurinae and Dicranurinae, 2 pairs in Notodontinae Subfamily: Thaumetopoeinae (1G 2S) Thaumetopoea (2S) 001 Thaumatopoiea processionea 002 Thaumatopoiea pityocampa (Oak Processionary) (Pine Processionary) fw: m14-16mm, f16-17mm; Jul-Sep; oaks Rare migrant with 2 British records 1966 & 2013 (Quercus spp); until 2006 a rare migrant, since then there have been outbreaks in London and Berkshire. See Forestry Commission (http://www.forestry.gov.uk/oakprocessionarymoth#outbreak stage) for more info, Subfamily: Cerurinae (2G 4S+1CI) Cerura (1S+1CI) 003 Cerura vinula (Puss Moth) 004 Cerura (Apocerura) erminea (Feline) 2 records from Jersey (/003-cerura-vinula-puss-moth.html) Furcula (3S) Antenna bipectinate, the pectinations shorter in female; labial palps very short, proboscis reduced; hindtibia with 1 pair of short spurs.