Download-The-Data (Accessed on 12 July 2021))
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Classification of Living and Fossil Genera of Decapod Crustaceans
RAFFLES BULLETIN OF ZOOLOGY 2009 Supplement No. 21: 1–109 Date of Publication: 15 Sep.2009 © National University of Singapore A CLASSIFICATION OF LIVING AND FOSSIL GENERA OF DECAPOD CRUSTACEANS Sammy De Grave1, N. Dean Pentcheff 2, Shane T. Ahyong3, Tin-Yam Chan4, Keith A. Crandall5, Peter C. Dworschak6, Darryl L. Felder7, Rodney M. Feldmann8, Charles H. J. M. Fransen9, Laura Y. D. Goulding1, Rafael Lemaitre10, Martyn E. Y. Low11, Joel W. Martin2, Peter K. L. Ng11, Carrie E. Schweitzer12, S. H. Tan11, Dale Tshudy13, Regina Wetzer2 1Oxford University Museum of Natural History, Parks Road, Oxford, OX1 3PW, United Kingdom [email protected] [email protected] 2Natural History Museum of Los Angeles County, 900 Exposition Blvd., Los Angeles, CA 90007 United States of America [email protected] [email protected] [email protected] 3Marine Biodiversity and Biosecurity, NIWA, Private Bag 14901, Kilbirnie Wellington, New Zealand [email protected] 4Institute of Marine Biology, National Taiwan Ocean University, Keelung 20224, Taiwan, Republic of China [email protected] 5Department of Biology and Monte L. Bean Life Science Museum, Brigham Young University, Provo, UT 84602 United States of America [email protected] 6Dritte Zoologische Abteilung, Naturhistorisches Museum, Wien, Austria [email protected] 7Department of Biology, University of Louisiana, Lafayette, LA 70504 United States of America [email protected] 8Department of Geology, Kent State University, Kent, OH 44242 United States of America [email protected] 9Nationaal Natuurhistorisch Museum, P. O. Box 9517, 2300 RA Leiden, The Netherlands [email protected] 10Invertebrate Zoology, Smithsonian Institution, National Museum of Natural History, 10th and Constitution Avenue, Washington, DC 20560 United States of America [email protected] 11Department of Biological Sciences, National University of Singapore, Science Drive 4, Singapore 117543 [email protected] [email protected] [email protected] 12Department of Geology, Kent State University Stark Campus, 6000 Frank Ave. -

New Zealand Oceanographic Institute Memoir 100
ISSN 0083-7903, 100 (Print) ISSN 2538-1016; 100 (Online) , , II COVER PHOTO. Dictyodendrilla cf. cavernosa (Lendenfeld, 1883) (type species of Dictyodendri/la Bergquist, 1980) (see page 24), from NZOI Stn I827, near Rikoriko Cave entrance, Poor Knights Islands Marine Reserve. Photo: Ken Grange, NZOI. This work is licensed under the Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/ NATIONAL INSTITUTE OF WATER AND ATMOSPHERIC RESEARCH The Marine Fauna of New Zealand: Index to the Fauna 2. Porifera by ELLIOT W. DAWSON N .Z. Oceanographic Institute, Wellington New Zealand Oceanographic Institute Memoir 100 1993 • This work is licensed under the Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/ Cataloguing in publication DAWSON, E.W. The marine fauna of New Zealand: Index to the Fauna 2. Porifera / by Elliot W. Dawson - Wellington: New Zealand Oceanographic Institute, 1993. (New Zealand Oceanographic Institute memoir, ISSN 0083-7903, 100) ISBN 0-478-08310-6 I. Title II. Series UDC Series Editor Dennis P. Gordon Typeset by Rose-Marie C. Thompson NIWA Oceanographic (NZOI) National Institute of Water and Atmospheric Research Received for publication: 17 July 1991 © NIWA Copyright 1993 2 This work is licensed under the Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/ CONTENTS Page ABSTRACT 5 INTRODUCTION 5 SCOPE AND ARRANGEMENT 7 SYSTEMATIC LIST 8 Class DEMOSPONGIAE 8 Subclass Homosclcromorpha .............................................................................................. -

Upogebia Deltaura (Crustacea: Thalassinidea) in Clyde Sea Maerl
BULLETIN OF MARINE SCIENCE, 58(3): 709-729, 1996 THE GENUS PARAXIOPSIS DE MAN, WITH DESCRIPTIONS OF NEW SPECIES FROM THE WESTERN ATLANTIC (CRUSTACEA: DECAPODA: AXIIDAE) Brian Kensley ABSTRACT The axiid shrimp genus Paraxiopsis De Man is reinstated, and separated from Eutricho- cheles Wood Mason on the basis of five characters. Nine species are assigned to Paraxiopsis, two from the Pacific and seven from the western Atlantic. Five of the latter are previously undescribed: P. foveolata from the eastern Gulf of Mexico; P. gracilimana from South Car olina to Tobaga, Belize, and the Gulf of Mexico; P. granulimana from Trinidad and the Florida Shelf; P. hispida from the Yucatan; and P. spinipleura from Belize and the Florida Keys. The type species, P. brocki De Man from the Indo-Pacific is redescribed, as is P. defensus Rathbun from the Caribbean. The species range from the intertidal to 95 m, in a variety of habitats including coral buttresses, coarse rubble, sand and mud. Axiid shrimps are, almost without exception, cryptic in habit. Consequently, little is known of their biology, while the taxa are often poorly represented in museum collections. Excluding the five new species described here, 14 species of axiids have been recorded in the tropical western Atlantic, of which the four species of Eiconaxius Bate, 1888, are all deepwater sponge commensals. Only Coralaxius nodulosus (Meinert, 1877) and Axiopsis serratifrons (A. Milne Ed wards, 1873) have been observed alive, and even for these species there is only scant biological information. The material described here has been accumulated over the course of several years, often incidental to collecting efforts for fishes and other crustacean groups, or as part of surveys of the shallow continental shelf. -

Taxonomy and Diversity of the Sponge Fauna from Walters Shoal, a Shallow Seamount in the Western Indian Ocean Region
Taxonomy and diversity of the sponge fauna from Walters Shoal, a shallow seamount in the Western Indian Ocean region By Robyn Pauline Payne A thesis submitted in partial fulfilment of the requirements for the degree of Magister Scientiae in the Department of Biodiversity and Conservation Biology, University of the Western Cape. Supervisors: Dr Toufiek Samaai Prof. Mark J. Gibbons Dr Wayne K. Florence The financial assistance of the National Research Foundation (NRF) towards this research is hereby acknowledged. Opinions expressed and conclusions arrived at, are those of the author and are not necessarily to be attributed to the NRF. December 2015 Taxonomy and diversity of the sponge fauna from Walters Shoal, a shallow seamount in the Western Indian Ocean region Robyn Pauline Payne Keywords Indian Ocean Seamount Walters Shoal Sponges Taxonomy Systematics Diversity Biogeography ii Abstract Taxonomy and diversity of the sponge fauna from Walters Shoal, a shallow seamount in the Western Indian Ocean region R. P. Payne MSc Thesis, Department of Biodiversity and Conservation Biology, University of the Western Cape. Seamounts are poorly understood ubiquitous undersea features, with less than 4% sampled for scientific purposes globally. Consequently, the fauna associated with seamounts in the Indian Ocean remains largely unknown, with less than 300 species recorded. One such feature within this region is Walters Shoal, a shallow seamount located on the South Madagascar Ridge, which is situated approximately 400 nautical miles south of Madagascar and 600 nautical miles east of South Africa. Even though it penetrates the euphotic zone (summit is 15 m below the sea surface) and is protected by the Southern Indian Ocean Deep- Sea Fishers Association, there is a paucity of biodiversity and oceanographic data. -

Decapod Crustacean Grooming: Functional Morphology, Adaptive Value, and Phylogenetic Significance
Decapod crustacean grooming: Functional morphology, adaptive value, and phylogenetic significance N RAYMOND T.BAUER Center for Crustacean Research, University of Southwestern Louisiana, USA ABSTRACT Grooming behavior is well developed in many decapod crustaceans. Antennular grooming by the third maxillipedes is found throughout the Decapoda. Gill cleaning mechanisms are qaite variable: chelipede brushes, setiferous epipods, epipod-setobranch systems. However, microstructure of gill cleaning setae, which are equipped with digitate scale setules, is quite conservative. General body grooming, performed by serrate setal brushes on chelipedes and/or posterior pereiopods, is best developed in decapods at a natant grade of body morphology. Brachyuran crabs exhibit less body grooming and virtually no specialized body grooming structures. It is hypothesized that the fouling pressures for body grooming are more severe in natant than in replant decapods. Epizoic fouling, particularly microbial fouling, and sediment fouling have been shown r I m ans of amputation experiments to produce severe effects on olfactory hairs, gills, and i.icubated embryos within short lime periods. Grooming has been strongly suggested as an important factor in the coevolution of a rhizocephalan parasite and its anomuran host. The behavioral organization of grooming is poorly studied; the nature of stimuli promoting grooming is not understood. Grooming characters may contribute to an understanding of certain aspects of decapod phylogeny. The occurrence of specialized antennal grooming brushes in the Stenopodidea, Caridea, and Dendrobranchiata is probably not due to convergence; alternative hypotheses are proposed to explain the distribution of this grooming character. Gill cleaning and general body grooming characters support a thalassinidean origin of the Anomura; the hypothesis of brachyuran monophyly is supported by the conservative and unique gill-cleaning method of the group. -
W7192e19.Pdf
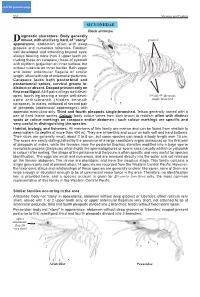
click for previous page 952 Shrimps and Prawns Sicyoniidae SICYONIIDAE Rock shrimps iagnostic characters: Body generally Drobust, with shell very hard, of “stony” grooves appearance; abdomen often with deep grooves and numerous tubercles. Rostrum well developed and extending beyond eyes, always bearing more than 3 upper teeth (in- cluding those on carapace); base of eyestalk with styliform projection on inner surface, but without tubercle on inner border. Both upper and lower antennular flagella of similar length, attached to tip of antennular peduncle. 1 Carapace lacks both postorbital and postantennal spines, cervical groove in- distinct or absent. Exopod present only on first maxilliped. All 5 pairs of legs well devel- 2 oped, fourth leg bearing a single well-devel- 3rd and 4th pleopods 4 single-branched oped arthrobranch (hidden beneath 3 carapace). In males, endopod of second pair 5 of pleopods (abdominal appendages) with appendix masculina only. Third and fourth pleopods single-branched. Telson generally armed with a pair of fixed lateral spines. Colour: body colour varies from dark brown to reddish; often with distinct spots or colour markings on carapace and/or abdomen - such colour markings are specific and very useful in distinguishing the species. Habitat, biology, and fisheries: All members of this family are marine and can be found from shallow to deep waters (to depths of more than 400 m). They are all benthic and occur on both soft and hard bottoms. Their sizes are generally small, about 2 to 8 cm, but some species can reach a body length over 15 cm. The sexes are easily distinguished by the presence of a large copulatory organ (petasma) on the first pair of pleopods of males, while the females have the posterior thoracic sternites modified into a large sperm receptacle process (thelycum) which holds the spermatophores or sperm sacs (usually whitish or yellowish in colour) after mating. -

The Genus Paraxiopsis De Man, with Descriptions of New Species from the Western Atlantic (Crustacea: Decapoda: Axiidae)
THE GENUS PARAXIOPSIS DE MAN, WITH DESCRIPTIONS OF NEW SPECIES FROM THE WESTERN ATLANTIC (CRUSTACEA: DECAPODA: AXIIDAE) The axiid shrimp genus Paraxiopsis De Man is reinstated, and separated from Eutricho- cheles Wood Mason on the basis of five characters. Nine species are assigned to Paraxiopsis, two from the Pacific and seven from the western Atlantic. Five of the latter are previously undescribed: P. foveolata from the eastern Gulf of Mexico; P. gracilimana from South Car- olina to Tobaga, Belize, and the Gulf of Mexico; P. granulimana from Trinidad and the Florida Shelf; P. hispida from the Yucatan; and P. spinipleura from Belize and the Florida Keys. The type species, P. brocki De Man from the Indo-Pacific is redescribed, as is P. defensus Rathbun from the Caribbean. The species range from the intertidal to 95 m, in a variety of habitats including coral buttresses, coarse rubble, sand and mud. Axiid shrimps are, almost without exception, cryptic in habit. Consequently, little is known of their biology, while the taxa are often poorly represented in museum collections. Excluding the five new species described here, 14 species ofaxiids have been recorded in the tropical western Atlantic, of which the four species of Eiconaxius Bate, 1888, are all deepwater sponge commensals. Only Coralaxius nodulosus (Meinert, 1877) and Axiopsis serratifrans (A. Milne Ed- wards, 1873) have been observed alive, and even for these species there is only scant biological information. The material described here has been accumulated over the course of several years, often incidental to collecting efforts for fishes and other crustacean groups, or as part of surveys of the shallow continental shelf. -

Prawn Fauna (Crustacea: Decapoda) of India - an Annotated Checklist of the Penaeoid, Sergestoid, Stenopodid and Caridean Prawns
Available online at: www.mbai.org.in doi: 10.6024/jmbai.2012.54.1.01697-08 Prawn fauna (Crustacea: Decapoda) of India - An annotated checklist of the Penaeoid, Sergestoid, Stenopodid and Caridean prawns E. V. Radhakrishnan*1, V. D. Deshmukh2, G. Maheswarudu3, Jose Josileen 1, A. P. Dineshbabu4, K. K. Philipose5, P. T. Sarada6, S. Lakshmi Pillai1, K. N. Saleela7, Rekhadevi Chakraborty1, Gyanaranjan Dash8, C.K. Sajeev1, P. Thirumilu9, B. Sridhara4, Y Muniyappa4, A.D.Sawant2, Narayan G Vaidya5, R. Dias Johny2, J. B. Verma3, P.K.Baby1, C. Unnikrishnan7, 10 11 11 1 7 N. P. Ramachandran , A. Vairamani , A. Palanichamy , M. Radhakrishnan and B. Raju 1CMFRI HQ, Cochin, 2Mumbai RC of CMFRI, 3Visakhapatnam RC of CMFRI, 4Mangalore RC of CMFRI, 5Karwar RC of CMFRI, 6Tuticorin RC of CMFRI, 7Vizhinjam RC of CMFRI, 8Veraval RC of CMFRI, 9Madras RC of CMFRI, 10Calicut RC of CMFRI, 11Mandapam RC of CMFRI *Correspondence e-mail: [email protected] Received: 07 Sep 2011, Accepted: 15 Mar 2012, Published: 30 Apr 2012 Original Article Abstract Many penaeoid prawns are of considerable value for the fishing Introduction industry and aquaculture operations. The annual estimated average landing of prawns from the fishery in India was 3.98 The prawn fauna inhabiting the marine, estuarine and lakh tonnes (2008-10) of which 60% were contributed by freshwater ecosystems of India are diverse and fairly well penaeid prawns. An additional 1.5 lakh tonnes is produced from known. Significant contributions to systematics of marine aquaculture. During 2010-11, India exported US $ 2.8 billion worth marine products, of which shrimp contributed 3.09% in prawns of Indian region were that of Milne Edwards (1837), volume and 69.5% in value of the total export. -

DEEP SEA LEBANON RESULTS of the 2016 EXPEDITION EXPLORING SUBMARINE CANYONS Towards Deep-Sea Conservation in Lebanon Project
DEEP SEA LEBANON RESULTS OF THE 2016 EXPEDITION EXPLORING SUBMARINE CANYONS Towards Deep-Sea Conservation in Lebanon Project March 2018 DEEP SEA LEBANON RESULTS OF THE 2016 EXPEDITION EXPLORING SUBMARINE CANYONS Towards Deep-Sea Conservation in Lebanon Project Citation: Aguilar, R., García, S., Perry, A.L., Alvarez, H., Blanco, J., Bitar, G. 2018. 2016 Deep-sea Lebanon Expedition: Exploring Submarine Canyons. Oceana, Madrid. 94 p. DOI: 10.31230/osf.io/34cb9 Based on an official request from Lebanon’s Ministry of Environment back in 2013, Oceana has planned and carried out an expedition to survey Lebanese deep-sea canyons and escarpments. Cover: Cerianthus membranaceus © OCEANA All photos are © OCEANA Index 06 Introduction 11 Methods 16 Results 44 Areas 12 Rov surveys 16 Habitat types 44 Tarablus/Batroun 14 Infaunal surveys 16 Coralligenous habitat 44 Jounieh 14 Oceanographic and rhodolith/maërl 45 St. George beds measurements 46 Beirut 19 Sandy bottoms 15 Data analyses 46 Sayniq 15 Collaborations 20 Sandy-muddy bottoms 20 Rocky bottoms 22 Canyon heads 22 Bathyal muds 24 Species 27 Fishes 29 Crustaceans 30 Echinoderms 31 Cnidarians 36 Sponges 38 Molluscs 40 Bryozoans 40 Brachiopods 42 Tunicates 42 Annelids 42 Foraminifera 42 Algae | Deep sea Lebanon OCEANA 47 Human 50 Discussion and 68 Annex 1 85 Annex 2 impacts conclusions 68 Table A1. List of 85 Methodology for 47 Marine litter 51 Main expedition species identified assesing relative 49 Fisheries findings 84 Table A2. List conservation interest of 49 Other observations 52 Key community of threatened types and their species identified survey areas ecological importanc 84 Figure A1. -

And Their Pa Laeoe Co Logical Significance
LATE CRETACEOUS SILICEOUS SPONGES FROM THE MIDDLE VISTULA RIVER VALLEY (CENTRAL POLAND) AND THEIR PA LAEOE CO LOGICAL SIGNIFICANCE Ewa ŚWIERCZEWSKA-GŁADYSZ Geological Department o f the Łódź University, Narutowicza 88, 90-139 Łódź, Poland; e-mail: [email protected] Świerczewska-Gładysz, E., 2006. Late Cretaceous siliceous sponges from the Middle Vistula River Valley (Central Poland) and their palaeoecological significance. Annales Societatis Geologorum Poloniae, 76: 227-296. Abstract: Siliceous sponges are extremely abundant in the Upper Campanian-Maastrichtian opokas and marls of the Middle Vis-ula River VaUey, situated in the western edge of the Lublin Basin, part of the Cre-aceous German-Polish Basin. This is also the only one area in Poland where strata bearing the Late Maastrichtian sponges are exposed. The presented paper is a taxonomic revision of sponges coUected from this region. Based both on existing and newly collected material comprising ca. 1750 specimens, 51 species have been described, including 18 belonging to the Hexactinosida, 15 - to the Lychniscosida and 18 - to Demospongiae. Among them, 28 have not been so far described from Poland. One new genus Varioporospongia, assigned to the family Ventriculitidae Smith and two new species Varioporospongia dariae sp. n. and Aphrocallistes calciformis sp. n. have been described. Comparison of sponge fauna from the area of Podilia, Crimea, Chernihov, and Donbas regions, as well as literature data point to the occurrence of species common in the analysed area and to the basins of Eastern and Western Europe. This in turn indicates good connections between particular basins of the European epicontinental sea dumg the Campanian-Maastrichtian. -

Hexasterophoran Glass Sponges of New Zealand (Porifera: Hexactinellida: Hexasterophora): Orders Hexactinosida, Aulocalycoida and Lychniscosida
Hexactinellida: Hexasterophora): Orders Hexactinosida, Aulocalycoida and Lychniscosida Aulocalycoida and Lychniscosida Hexactinellida: Hexasterophora): Orders Hexactinosida, The Marine Fauna of New Zealand: Hexasterophoran Glass Sponges Zealand (Porifera: ISSN 1174–0043; 124 Henry M. Reiswig and Michelle Kelly The Marine Fauna of New Zealand: Hexasterophoran Glass Sponges of New Zealand (Porifera: Hexactinellida: Hexasterophora): Orders Hexactinosida, Aulocalycoida and Lychniscosida Henry M. Reiswig and Michelle Kelly NIWA Biodiversity Memoir 124 COVER PHOTO Two unidentified hexasterophoran glass sponge species, the first possibly Farrea onychohexastera n. sp. (frilly white honeycomb sponge in several bushy patches), and the second possibly Chonelasma lamella, but also possibly C. chathamense n. sp. (lower left white fan), attached to the habitat-forming coral Solenosmilia variabilis, dominant at 1078 m on the Graveyard seamount complex of the Chatham Rise (NIWA station TAN0905/29: 42.726° S, 179.897° W). Image captured by DTIS (Deep Towed Imaging System) onboard RV Tangaroa, courtesy of NIWA Seamounts Programme (SFAS103), Oceans2020 (LINZ, MFish) and Rob Stewart, NIWA, Wellington (Photo: NIWA). This work is licensed under the Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/ NATIONAL INSTITUTE OF WATER AND ATMOSPHERIC RESEARCH (NIWA) The Marine Fauna of New Zealand: Hexasterophoran Glass Sponges of New Zealand (Porifera: Hexactinellida: -

<I>Bartholomea Annulata</I>
BULLETIN OF MARINE SCIENCE, 37(3): 893-904,1985 CORAL REEF PAPER TWO MORE SIBLING SPECIES OF ALPHEID SHRIMPS ASSOCIATED WITH THE CARIBBEAN SEA ANEMONES BARTHOLOMEA ANNULATA AND HETERACTIS LUCIDA Nancy Knowlton and Brian D. Keller ABSTRACT We have described two new species of snapping shrimp, Alpheus polystictus and A. ro- quensis. The new species form part of a complex of four sibling species associated with Caribbean sea anemones, the others being the well-known A. armatus Rathbun, 1900 and the recently describedA. immaculatus Knowlton and Keller, 1983. Alpheus roquensis is found with the anemone Heteractis lucida. while the other three shrimps live with Bartholomea annulata. In laboratory choice experiments, each shrimp species prefers the species of an em- one with which it is typically found in the field, although each can shelter under the other species of anemone. All four species are extremely similar morphologically, being distin- guished largely on the basis of color pattern. The validity of the species is confirmed by the total absence of interbreeding; heterospecific male-female pairs are never found in the field, and it is impossible to force pairings between species in the laboratory. Alpheus polystictus is rare in Jamaica and Haiti, while in Venezuela it is sometimes the dominant species to depths of 10 m. In the areas examined, it has always occurred with at least one of the other two Bartholomea associates. The geographic distribution of A. roquensis is more limited, as there are no reports of alpheids associated with Heteractis lucida, and none has been found with this anemone in Jamaica.