Molecular Phylogeny of Hingebeak Shrimps
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Classification of Living and Fossil Genera of Decapod Crustaceans
RAFFLES BULLETIN OF ZOOLOGY 2009 Supplement No. 21: 1–109 Date of Publication: 15 Sep.2009 © National University of Singapore A CLASSIFICATION OF LIVING AND FOSSIL GENERA OF DECAPOD CRUSTACEANS Sammy De Grave1, N. Dean Pentcheff 2, Shane T. Ahyong3, Tin-Yam Chan4, Keith A. Crandall5, Peter C. Dworschak6, Darryl L. Felder7, Rodney M. Feldmann8, Charles H. J. M. Fransen9, Laura Y. D. Goulding1, Rafael Lemaitre10, Martyn E. Y. Low11, Joel W. Martin2, Peter K. L. Ng11, Carrie E. Schweitzer12, S. H. Tan11, Dale Tshudy13, Regina Wetzer2 1Oxford University Museum of Natural History, Parks Road, Oxford, OX1 3PW, United Kingdom [email protected] [email protected] 2Natural History Museum of Los Angeles County, 900 Exposition Blvd., Los Angeles, CA 90007 United States of America [email protected] [email protected] [email protected] 3Marine Biodiversity and Biosecurity, NIWA, Private Bag 14901, Kilbirnie Wellington, New Zealand [email protected] 4Institute of Marine Biology, National Taiwan Ocean University, Keelung 20224, Taiwan, Republic of China [email protected] 5Department of Biology and Monte L. Bean Life Science Museum, Brigham Young University, Provo, UT 84602 United States of America [email protected] 6Dritte Zoologische Abteilung, Naturhistorisches Museum, Wien, Austria [email protected] 7Department of Biology, University of Louisiana, Lafayette, LA 70504 United States of America [email protected] 8Department of Geology, Kent State University, Kent, OH 44242 United States of America [email protected] 9Nationaal Natuurhistorisch Museum, P. O. Box 9517, 2300 RA Leiden, The Netherlands [email protected] 10Invertebrate Zoology, Smithsonian Institution, National Museum of Natural History, 10th and Constitution Avenue, Washington, DC 20560 United States of America [email protected] 11Department of Biological Sciences, National University of Singapore, Science Drive 4, Singapore 117543 [email protected] [email protected] [email protected] 12Department of Geology, Kent State University Stark Campus, 6000 Frank Ave. -

Thais Peixoto Macedo DE LIMPADORES a ORNAMENTOS DE AQUÁRIO: a Diversidade De Camarões Recifais Em Unidades De Conservação D
Thais Peixoto Macedo DE LIMPADORES A ORNAMENTOS DE AQUÁRIO: A diversidade de camarões recifais em Unidades de Conservação da costa brasileira Trabalho de Conclusão de Curso apresentado ao programa de graduação do Curso de Ciências Biológicas da Universidade Federal de Santa Catarina em cumprimento a requisito parcial para a obtenção do grau de bacharel em Ciências Biológicas. Orientadora: Profa. Dra. Andrea Santarosa Freire Florianópolis 2018 Ficha de identificação da obra elaborada pelo autor através do Programa de Geração Automática da Biblioteca Universitária da UFSC. Macedo, Thais Peixoto DE LIMPADORES A ORNAMENTOS DE AQUÁRIO : A diversidade de camarões recifais em Unidades de Conservação da costa brasileira / Thais Peixoto Macedo ; orientadora, Andrea Santarosa Freire, 2018. 65 p. Trabalho de Conclusão de Curso (graduação) - Universidade Federal de Santa Catarina, Centro de Ciências Biológicas, Graduação em Ciências Biológicas, Florianópolis, 2018. Inclui referências. 1. Ciências Biológicas. 2. Diversidade taxonômica. 3. Padrões de diversidade. 4. Lista de espécies. 5. Caridea e Stenopodidea. I. Freire, Andrea Santarosa. II. Universidade Federal de Santa Catarina. Graduação em Ciências Biológicas. III. Título. Thais Peixoto Macedo DE LIMPADORES A ORNAMENTOS DE AQUÁRIO: A diversidade de camarões recifais em Unidades de Conservação da costa brasileira Este Trabalho de Conclusão de Curso foi julgado adequado para obtenção do Título de “Bacharel em Ciências Biológicas” e aprovada em sua forma final pela Universidade Federal de Santa Catarina Florianópolis, 6 de dezembro de 2018. ________________________ Prof. Dr. Carlos Zanetti Coordenador do Curso Banca Examinadora: ________________________ Prof.ª Dr.ª Andrea Santarosa Freire Orientadora Universidade Federal de Santa Catarina ________________________ Prof. Dr. Sergio Floeter Universidade Federal de Santa Catarina ________________________ Tammy Arai Iwasa Universidade Estadual de Campinas AGRADECIMENTOS Ciência não se faz sozinho. -

Diversity of Seagrass-Associated Decapod Crustaceans in a Tropical Reef Lagoon Prior to Large Environmental Changes: a Baseline Study
diversity Article Diversity of Seagrass-Associated Decapod Crustaceans in a Tropical Reef Lagoon Prior to Large Environmental Changes: A Baseline Study Patricia Briones-Fourzán * , Luz Verónica Monroy-Velázquez, Jaime Estrada-Olivo y and Enrique Lozano-Álvarez Unidad Académica de Sistemas Arrecifales, Instituto de Ciencias del Mar y Limnología, Universidad Nacional Autónoma de Mexico, Puerto Morelos, 77580 Quintana Roo, Mexico; [email protected] (L.V.M.-V.); [email protected] (J.E.-O.); [email protected] (E.L.-Á.) * Correspondence: [email protected] Current address: Calle Trasatlántico SM-18, Mz 24, Lote 12; Villas Morelos, Puerto Morelos, y 77580 Quintana Roo, Mexico. Received: 7 April 2020; Accepted: 19 May 2020; Published: 23 May 2020 Abstract: The community composition of decapods associated with subtidal tropical seagrass meadows was analyzed in a pristine reef lagoon on the Mexican Caribbean coast in the summer of 1995 and winter of 1998. The macrophyte community was dominated by Thalassia testudinum followed by Syringodium filiforme, with interspersed rhyzophytic macroalgae and large patches of drift algae. In each season, 10 one-min trawls were made with an epibenthic sled (mesh aperture 1 mm) during the day and 10 during the night on each of five sites. In all, 53,211 decapods belonging to 119 species were collected. The most diverse taxa were Brachyura and Caridea, but the most abundant were Caridea and Anomura. Dominance was high, with three species (Latreutes fucorum, Cuapetes americanus, and Thor manningi) accounting for almost 50% of individuals, and 10 species accounting for nearly 90% of individuals. There was great similarity in community composition and ecological indices between seasons, but significantly more individuals and species in night versus day samples. -
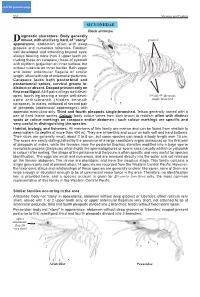
W7192e19.Pdf
click for previous page 952 Shrimps and Prawns Sicyoniidae SICYONIIDAE Rock shrimps iagnostic characters: Body generally Drobust, with shell very hard, of “stony” grooves appearance; abdomen often with deep grooves and numerous tubercles. Rostrum well developed and extending beyond eyes, always bearing more than 3 upper teeth (in- cluding those on carapace); base of eyestalk with styliform projection on inner surface, but without tubercle on inner border. Both upper and lower antennular flagella of similar length, attached to tip of antennular peduncle. 1 Carapace lacks both postorbital and postantennal spines, cervical groove in- distinct or absent. Exopod present only on first maxilliped. All 5 pairs of legs well devel- 2 oped, fourth leg bearing a single well-devel- 3rd and 4th pleopods 4 single-branched oped arthrobranch (hidden beneath 3 carapace). In males, endopod of second pair 5 of pleopods (abdominal appendages) with appendix masculina only. Third and fourth pleopods single-branched. Telson generally armed with a pair of fixed lateral spines. Colour: body colour varies from dark brown to reddish; often with distinct spots or colour markings on carapace and/or abdomen - such colour markings are specific and very useful in distinguishing the species. Habitat, biology, and fisheries: All members of this family are marine and can be found from shallow to deep waters (to depths of more than 400 m). They are all benthic and occur on both soft and hard bottoms. Their sizes are generally small, about 2 to 8 cm, but some species can reach a body length over 15 cm. The sexes are easily distinguished by the presence of a large copulatory organ (petasma) on the first pair of pleopods of males, while the females have the posterior thoracic sternites modified into a large sperm receptacle process (thelycum) which holds the spermatophores or sperm sacs (usually whitish or yellowish in colour) after mating. -

Lysmata Amboinensis (De Man, 1888)
Lysmata amboinensis (de Man, 1888) B. Santhosh, M. K. Anil and Biji Xavier IDENTIFICATION Order : Decapoda Family : Lysmatidae Common/FAO : Pacific cleaner Name (English) shrimp Local names:names Not available MORPHOLOGICAL DESCRIPTION The Pacific cleaner shrimp is easily identified by its colour patterns. The body is light brown with one white band dorsally and two red bands laterally running longitudinally. The tail has two white spots on either side. The antennae are white in colour and the first pair has red coloured base. It grows up to a maximum of 6 cm. Source of image : RC CMFRI, Vizhinjam 363 PROFILE GEOGRAPHICAL DISTRIBUTION Scarlet cleaner shrimp or Pacific cleaner shrimp is one of the most popular species of ornamental crustaceans distributed in the waters of the Indo-Pacific region in Indonesia and Sri Lanka. HABITAT AND BIOLOGY It is one of the popular marine shrimp, associated with coral reefs and compatible with smaller sized marine ornamental fishes. It hides in the near shore, shallow and protected areas within a temperature range of 25-30 °C. In the Indo-Pacific areas and the Red Sea, it is mostly found in caves and crevices of coral reefs. It especially needs shelter from predators when it is moulting. It is an omnivore and a scavenger and often feeds on the external parasites of fishes. As its name indicates, this species cleans fishes including moray eels and groupers feeding on their external parasites as well as on mucous and dead or injured tissue. The shrimp moults once every 3-8 weeks and spawns regularly every 2-3 weeks. -

From the Persian Gulf
ZOBODAT - www.zobodat.at Zoologisch-Botanische Datenbank/Zoological-Botanical Database Digitale Literatur/Digital Literature Zeitschrift/Journal: Annalen des Naturhistorischen Museums in Wien Jahr/Year: 2007 Band/Volume: 108B Autor(en)/Author(s): De Grave Sammy Artikel/Article: Notes on some shrimp species (Decapoda: Caridea) from the Persian Gulf. 145-152 ©Naturhistorisches Museum Wien, download unter www.biologiezentrum.at Ann. Naturhist. Mus. Wien 108 B 145- 152 Wien, Mai 2007 Notes on some shrimp species (Decapoda: Caridea) from the Persian Gulf S. DE GRAVE* Abstract A report is presented on a small collection of caridean shrimp (Crustacea: Decapoda) from coastal waters of the United Arab Emirates in the Persian Gulf. Eight species are new records for the area, raising the total number of carideans known from the Persian Gulf to 46. A review is presented of all previous records, which highlights the relative paucity of records. Key words: Decapoda, Caridea, Persian Gulf, new records Zusammenfassung Diese Arbeit behandelt eine kleine Sammlung von Garnelen aus den Küstengewässern der Vereinigten Arabischen Emirate im Persischen Golf. Acht Arten werden zum ersten Mal aus diesem Gebiet gemeldet, das erhöht die Gesamtzahl der aus dem Golf bekannten Caridea auf 46. Eine Übersicht aller bisherigen Funde zeigt auf wie wenig aus diesem Gebiet vorliegt. Introduction NOBILI (1905a, b) described four species of caridean shrimp from the Persian Gulf: Alpheus bucephaloides NOBILI, 1905; Alpheuspersicus NOBILI, 1905 [now considered a junior synonym of Alpheus malleodigitus (BATE, 1888)]; Periclimenes borradailei NOBILI, 1905; and Harpilius gerlacheiNoBiu, 1905 (now Philarius gerlachei). In 1906, Nobili in a major review of the material collected by J. -

Understanding Transformative Forces of Aquaculture in the Marine Aquarium Trade
The University of Maine DigitalCommons@UMaine Electronic Theses and Dissertations Fogler Library Summer 8-22-2020 Senders, Receivers, and Spillover Dynamics: Understanding Transformative Forces of Aquaculture in the Marine Aquarium Trade Bryce Risley University of Maine, [email protected] Follow this and additional works at: https://digitalcommons.library.umaine.edu/etd Part of the Marine Biology Commons Recommended Citation Risley, Bryce, "Senders, Receivers, and Spillover Dynamics: Understanding Transformative Forces of Aquaculture in the Marine Aquarium Trade" (2020). Electronic Theses and Dissertations. 3314. https://digitalcommons.library.umaine.edu/etd/3314 This Open-Access Thesis is brought to you for free and open access by DigitalCommons@UMaine. It has been accepted for inclusion in Electronic Theses and Dissertations by an authorized administrator of DigitalCommons@UMaine. For more information, please contact [email protected]. SENDERS, RECEIVERS, AND SPILLOVER DYNAMICS: UNDERSTANDING TRANSFORMATIVE FORCES OF AQUACULTURE IN THE MARINE AQUARIUM TRADE By Bryce Risley B.S. University of New Mexico, 2014 A THESIS Submitted in Partial Fulfillment of the Requirements for the Degree of Master of Science (in Marine Policy and Marine Biology) The Graduate School The University of Maine May 2020 Advisory Committee: Joshua Stoll, Assistant Professor of Marine Policy, Co-advisor Nishad Jayasundara, Assistant Professor of Marine Biology, Co-advisor Aaron Strong, Assistant Professor of Environmental Studies (Hamilton College) Christine Beitl, Associate Professor of Anthropology Douglas Rasher, Senior Research Scientist of Marine Ecology (Bigelow Laboratory) Heather Hamlin, Associate Professor of Marine Biology No photograph in this thesis may be used in another work without written permission from the photographer. -

Prawn Fauna (Crustacea: Decapoda) of India - an Annotated Checklist of the Penaeoid, Sergestoid, Stenopodid and Caridean Prawns
Available online at: www.mbai.org.in doi: 10.6024/jmbai.2012.54.1.01697-08 Prawn fauna (Crustacea: Decapoda) of India - An annotated checklist of the Penaeoid, Sergestoid, Stenopodid and Caridean prawns E. V. Radhakrishnan*1, V. D. Deshmukh2, G. Maheswarudu3, Jose Josileen 1, A. P. Dineshbabu4, K. K. Philipose5, P. T. Sarada6, S. Lakshmi Pillai1, K. N. Saleela7, Rekhadevi Chakraborty1, Gyanaranjan Dash8, C.K. Sajeev1, P. Thirumilu9, B. Sridhara4, Y Muniyappa4, A.D.Sawant2, Narayan G Vaidya5, R. Dias Johny2, J. B. Verma3, P.K.Baby1, C. Unnikrishnan7, 10 11 11 1 7 N. P. Ramachandran , A. Vairamani , A. Palanichamy , M. Radhakrishnan and B. Raju 1CMFRI HQ, Cochin, 2Mumbai RC of CMFRI, 3Visakhapatnam RC of CMFRI, 4Mangalore RC of CMFRI, 5Karwar RC of CMFRI, 6Tuticorin RC of CMFRI, 7Vizhinjam RC of CMFRI, 8Veraval RC of CMFRI, 9Madras RC of CMFRI, 10Calicut RC of CMFRI, 11Mandapam RC of CMFRI *Correspondence e-mail: [email protected] Received: 07 Sep 2011, Accepted: 15 Mar 2012, Published: 30 Apr 2012 Original Article Abstract Many penaeoid prawns are of considerable value for the fishing Introduction industry and aquaculture operations. The annual estimated average landing of prawns from the fishery in India was 3.98 The prawn fauna inhabiting the marine, estuarine and lakh tonnes (2008-10) of which 60% were contributed by freshwater ecosystems of India are diverse and fairly well penaeid prawns. An additional 1.5 lakh tonnes is produced from known. Significant contributions to systematics of marine aquaculture. During 2010-11, India exported US $ 2.8 billion worth marine products, of which shrimp contributed 3.09% in prawns of Indian region were that of Milne Edwards (1837), volume and 69.5% in value of the total export. -

The Complete Mitogenome of €Lysmata Vittata
bioRxiv preprint doi: https://doi.org/10.1101/2021.08.04.455109; this version posted August 5, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 The complete mitogenome of Lysmata vittata (Crustacea: 2 Decapoda: Hippolytidae) and its phylogenetic position in 3 Decapoda 4 Longqiang Zhu1,2,3, Zhihuang Zhu1,2*, Leiyu Zhu1,2, Dingquan Wang3, Jianxin 5 Wang3*, Qi Lin1,2,3* 6 1 Fisheries Research Institute of Fujian, Xiamen, China 7 2 Key Laboratory of Cultivation and High-value Utilization of Marine Organisms in Fujian Province, Xiamen, 8 China 9 3Marine Microorganism Ecological & Application Lab, Zhejiang Ocean University, Zhejiang, China 10 * [email protected] (QL); [email protected] (JW); [email protected] (ZZ) 11 12 Abstract 13 In this study, the complete mitogenome of Lysmata vittata (Crustacea: Decapoda: 14 Hippolytidae) has been determined. The genome sequence was 22003 base pairs (bp) 15 and it included thirteen protein-coding genes (PCGs), twenty-two transfer RNA genes 16 (tRNAs), two ribosomal RNA genes (rRNAs) and three putative control regions 17 (CRs). The nucleotide composition of AT was 71.50%, with a slightly negative AT 18 skewness (-0.04). Usually the standard start codon of the PCGs was ATN, while cox1, 19 nad4L and cox3 began with TTG, TTG and GTG. The canonical termination codon 20 was TAA, while nad5 and nad4 ended with incomplete stop codon T, and cox1 ended 21 with TAG. -

Download-The-Data (Accessed on 12 July 2021))
diversity Article Integrative Taxonomy of New Zealand Stenopodidea (Crustacea: Decapoda) with New Species and Records for the Region Kareen E. Schnabel 1,* , Qi Kou 2,3 and Peng Xu 4 1 Coasts and Oceans Centre, National Institute of Water & Atmospheric Research, Private Bag 14901 Kilbirnie, Wellington 6241, New Zealand 2 Institute of Oceanology, Chinese Academy of Sciences, Qingdao 266071, China; [email protected] 3 College of Marine Science, University of Chinese Academy of Sciences, Beijing 100049, China 4 Key Laboratory of Marine Ecosystem Dynamics, Second Institute of Oceanography, Ministry of Natural Resources, Hangzhou 310012, China; [email protected] * Correspondence: [email protected]; Tel.: +64-4-386-0862 Abstract: The New Zealand fauna of the crustacean infraorder Stenopodidea, the coral and sponge shrimps, is reviewed using both classical taxonomic and molecular tools. In addition to the three species so far recorded in the region, we report Spongicola goyi for the first time, and formally describe three new species of Spongicolidae. Following the morphological review and DNA sequencing of type specimens, we propose the synonymy of Spongiocaris yaldwyni with S. neocaledonensis and review a proposed broad Indo-West Pacific distribution range of Spongicoloides novaezelandiae. New records for the latter at nearly 54◦ South on the Macquarie Ridge provide the southernmost record for stenopodidean shrimp known to date. Citation: Schnabel, K.E.; Kou, Q.; Xu, Keywords: sponge shrimp; coral cleaner shrimp; taxonomy; cytochrome oxidase 1; 16S ribosomal P. Integrative Taxonomy of New RNA; association; southwest Pacific Ocean Zealand Stenopodidea (Crustacea: Decapoda) with New Species and Records for the Region. -

Espécies Marinhas De Cabo Verde (BIOTECMAR)
EQUINODERMES 77 Ophiothrix fragilis OfiuróideOfiuríde-comum Glatter SchlangensternZerbrechlicher Schlangenstern OfiuraOfiura fina Ophiure lisse Ophiure fragile AnnulatedBrittle star brittle star Stella serpentina Stellaliscia serpentina spinosa Ophioderma longicaudum Echinometra lucunter Nieves González Ofiuróide Ouriço-preto-de-poças Glatter Schlangenstern Felsenbohrerseeigel Ofiura Erizo de mar Ophiure lisse Oursin de récif Annulated brittle star Rock boring urchin Stella serpentina liscia Riccio de mare 78 Diadema africanum Centrostephanus longispinus Maite Vázquez Ouriço-de-espinhos-compridos Diademseeigel Ouriço-de-espinhos-compridos Diademseeigel Diadema, eriza Oursin diadème africain Puerco espín marino * Oursin diadème méditerranéen * Long-spined black sea urchin Riccio diadema Largest urchin Riccio diadema Eucidaris tribuloides Holothuria surinamensis Oriço, ouriço Pon-di-mar, biroti Ostatlantik-Lanzenseeigel Variable Seegurke Erizo de palitos Oursin-lance antillais Holoturia, pepino de mar Concombre de mer Slate pen sea urchin Riccio matita Variable sea cucumber Cetriolo di mare punte nere EQUINODERMES 79 Holothuria lentiginosa Isostichopus badionotus Pon d´mar, biroti Pepino-do-mar Seegurke Sommersprossen Seegurke Pepino de pecas Concombre de mer tacheté Pepino de mar Holothurie à points Sea cucumber freckles Cetriolo di mare lentigginoso Sea cucumber Cetriolo di mare Holothuria mammata Euapta lappa Wurmseegurke Pon d´mar, biroti Variable Seegurke Pepino-do-mar Holothurie-serpent collante Holoturia, pepino de mar Concombre cracheur -

Redalyc.Larval Development of the Rock Shrimp Rhynchocinetes Typus
Revista Chilena de Historia Natural ISSN: 0716-078X [email protected] Sociedad de Biología de Chile Chile DUPRÉ, ENRIQUE; FLORES, LUIS; PALMA, SERGIO Larval development of the rock shrimp Rhynchocinetes typus Milne Edwards, 1937 (Decapoda, Caridea) reared in the laboratory Revista Chilena de Historia Natural, vol. 81, núm. 2, 2008, pp. 155-170 Sociedad de Biología de Chile Santiago, Chile Available in: http://www.redalyc.org/articulo.oa?id=369944286001 How to cite Complete issue Scientific Information System More information about this article Network of Scientific Journals from Latin America, the Caribbean, Spain and Portugal Journal's homepage in redalyc.org Non-profit academic project, developed under the open access initiative LARVAL DEVELOPMENT OF RHYNCHOCINETES TYPUSRevista Chilena de Historia Natural155 81: 155-170, 2008 Larval development of the rock shrimp Rhynchocinetes typus Milne Edwards, 1937 (Decapoda, Caridea) reared in the laboratory Desarrollo larval del camarón de roca Rhynchocinetes typus Milne Edwards, 1937 (Decapoda, Caridea) cultivados en laboratorio ENRIQUE DUPRÉ1*, LUIS FLORES1 & SERGIO PALMA2 1 Departamento de Biología Marina, Facultad de Ciencias del Mar, Universidad Católica del Norte, Casilla117 Coquimbo, Chile; [email protected] 2 Escuela de Ciencias del Mar, Pontificia Universidad Católica de Valparaíso, Valparaíso, Chile *e-mail for correspondence: [email protected] ABSTRACT The first description of the larval stages of a representative of the family Rhynchocinetidae from the southeastern Pacific coast of South America is presented. Larvae of the rock shrimp Rhynchocinetes typus, from rocky subtidal environment of the Chile-Peru coast were reared in the laboratory at 22 ºC, salinity 32 and feeding with Artemia franciscana. Seven zoeal stages, which occur through 10 successive moults, are described and illustrated in detail.