GSDMB Induces an Asthma Phenotype Characterized by Increased Airway Responsiveness and Remodeling Without Lung Inflammation
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Original Article Tocilizumab Infusion Therapy Normalizes Inflammation in Sporadic ALS Patients
Am J Neurodegener Dis 2013;2(2):129-139 www.AJND.us /ISSN:2165-591X/AJND1304002 Original Article Tocilizumab infusion therapy normalizes inflammation in sporadic ALS patients Milan Fiala1, Mathew T Mizwicki1, Rachel Weitzman1, Larry Magpantay2, Norihiro Nishimoto3 1Department of Surgery, David Geffen School of Medicine at UCLA, 100 UCLA Medical Plaza, Suite 220, Los Angeles, CA 90095-6970, USA; 2Department of Obstetrics and Gynecology, David Geffen School of Medicine at UCLA, Los Angeles, 650 Charles E. Young Drive, Los Angeles, CA, 90095-1735, USA; 3Department of Molecular Regulation for Intractable Diseases, Institute of Medical Sciences, Tokyo Medical University, Minamisenba, Chuo- ku, Osaka, 542-0081, Japan Received April 8 2013; Accepted May 19 2013; Epub June 21, 2013; Published July 1, 2013 Abstract: Patients with sporadic amyotrophic lateral sclerosis (sALS) show inflammation in the spinal cord and pe- ripheral blood. The inflammation is driven by stimulation of macrophages by aggregated superoxide dismutase 1 (SOD1) through caspase1, interleukin 1 (IL1), IL6 and chemokine signaling. Inflammatory gene activation is inhibit- ed in vitro by tocilizumab, a humanized antibody to IL6 receptor (IL6R). Tocilizumab inhibits global interleukin-6 (IL6) signaling, a key mechanism in chronic rheumatoid disorders. Here we studied in vivo baseline inflammatory gene transcription in peripheral blood mononuclear cells (PBMCs) of 10 sALS patients, and the effects of tocilizumab (ActemraR) infusions. At baseline, one half of ALS subjects had strong inflammatory activation (Group 1) (8 genes up regulated >4-fold, P<0.05 vs. controls) and the other half (Group 2) had weak activation. All patients showed greater than four-fold up regulation of MMP1, CCL7, CCL13 and CCL24. -
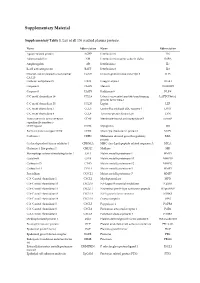
Supplementary Material
Supplementary Material Supplementary Table 1. List of all 136 studied plasma proteins. Name Abbreviation Name Abbreviation Agouti-related protein AGRP Interleukin-6 IL6 Adrenomedullin AM Interleukin-6 receptor subunit alpha IL6RA Amphiregulin AR Interleukin-7 IL7 B-cell activating factor BAFF Interleukin-8 IL8 Ovarian cancer-related tumor marker CA125 Immunoglobulin-like transcript 3 ILT3 CA 125 Carbonic anhydrase IX CAIX Integrin alpha-1 ITGA1 Caspase-3 CASP3 Melusin ITGB1BP2 Caspase-8 CASP8 Kallikrein-6 KLK6 C-C motif chemokine 19 CCL19 Latency-associated peptide transforming LAPTGFbeta1 growth factor beta-1 C-C motif chemokine 20 CCL20 Leptin LEP C-C motif chemokine 3 CCL3 Lectin-like oxidized LDL receptor 1 LOX1 C-C motif chemokine 4 CCL4 Tyrosine-protein kinase Lyn LYN Tumor necrosis factor receptor CD40 Membrane-bound aminopeptidase P mAmP superfamily member 5 CD40 ligand CD40L Myoglobin MB Early activation antigen CD69 CD69 Monocyte chemotactic protein 1 MCP1 Cadherin-3 CDH3 Melanoma-derived growth regulatory MIA protein Cyclin-dependent kinase inhibitor 1 CDKN1A MHC class I polypeptide-related sequence A MICA Chitinase-3-like protein 1 CHI3L1 Midkine MK Macrophage colony-stimulating factor 1 CSF1 Matrix metalloproteinase-1 MMP1 Cystatin-B CSTB Matrix metalloproteinase-10 MMP10 Cathepsin D CTSD Matrix metalloproteinase-12 MMP12 Cathepsin L1 CTSL1 Matrix metalloproteinase-3 MMP3 Fractalkine CX3CL1 Matrix metalloproteinase-7 MMP7 C-X-C motif chemokine 1 CXCL1 Myeloperoxidase MPO C-X-C motif chemokine 10 CXCL10 NF-kappa-B essential -

Integrating Single-Step GWAS and Bipartite Networks Reconstruction Provides Novel Insights Into Yearling Weight and Carcass Traits in Hanwoo Beef Cattle
animals Article Integrating Single-Step GWAS and Bipartite Networks Reconstruction Provides Novel Insights into Yearling Weight and Carcass Traits in Hanwoo Beef Cattle Masoumeh Naserkheil 1 , Abolfazl Bahrami 1 , Deukhwan Lee 2,* and Hossein Mehrban 3 1 Department of Animal Science, University College of Agriculture and Natural Resources, University of Tehran, Karaj 77871-31587, Iran; [email protected] (M.N.); [email protected] (A.B.) 2 Department of Animal Life and Environment Sciences, Hankyong National University, Jungang-ro 327, Anseong-si, Gyeonggi-do 17579, Korea 3 Department of Animal Science, Shahrekord University, Shahrekord 88186-34141, Iran; [email protected] * Correspondence: [email protected]; Tel.: +82-31-670-5091 Received: 25 August 2020; Accepted: 6 October 2020; Published: 9 October 2020 Simple Summary: Hanwoo is an indigenous cattle breed in Korea and popular for meat production owing to its rapid growth and high-quality meat. Its yearling weight and carcass traits (backfat thickness, carcass weight, eye muscle area, and marbling score) are economically important for the selection of young and proven bulls. In recent decades, the advent of high throughput genotyping technologies has made it possible to perform genome-wide association studies (GWAS) for the detection of genomic regions associated with traits of economic interest in different species. In this study, we conducted a weighted single-step genome-wide association study which combines all genotypes, phenotypes and pedigree data in one step (ssGBLUP). It allows for the use of all SNPs simultaneously along with all phenotypes from genotyped and ungenotyped animals. Our results revealed 33 relevant genomic regions related to the traits of interest. -

The Effect of Hypoxia on the Expression of CXC Chemokines and CXC Chemokine Receptors—A Review of Literature
International Journal of Molecular Sciences Review The Effect of Hypoxia on the Expression of CXC Chemokines and CXC Chemokine Receptors—A Review of Literature Jan Korbecki 1 , Klaudyna Kojder 2, Patrycja Kapczuk 1, Patrycja Kupnicka 1 , Barbara Gawro ´nska-Szklarz 3 , Izabela Gutowska 4 , Dariusz Chlubek 1 and Irena Baranowska-Bosiacka 1,* 1 Department of Biochemistry and Medical Chemistry, Pomeranian Medical University in Szczecin, Powsta´nców Wielkopolskich 72 Av., 70-111 Szczecin, Poland; [email protected] (J.K.); [email protected] (P.K.); [email protected] (P.K.); [email protected] (D.C.) 2 Department of Anaesthesiology and Intensive Care, Pomeranian Medical University in Szczecin, Unii Lubelskiej 1, 71-281 Szczecin, Poland; [email protected] 3 Department of Pharmacokinetics and Therapeutic Drug Monitoring, Pomeranian Medical University in Szczecin, Powsta´nców Wielkopolskich 72 Av., 70-111 Szczecin, Poland; [email protected] 4 Department of Medical Chemistry, Pomeranian Medical University in Szczecin, Powsta´nców Wlkp. 72 Av., 70-111 Szczecin, Poland; [email protected] * Correspondence: [email protected]; Tel.: +48-914661515 Abstract: Hypoxia is an integral component of the tumor microenvironment. Either as chronic or cycling hypoxia, it exerts a similar effect on cancer processes by activating hypoxia-inducible factor-1 (HIF-1) and nuclear factor (NF-κB), with cycling hypoxia showing a stronger proinflammatory influ- ence. One of the systems affected by hypoxia is the CXC chemokine system. This paper reviews all available information on hypoxia-induced changes in the expression of all CXC chemokines (CXCL1, CXCL2, CXCL3, CXCL4, CXCL5, CXCL6, CXCL7, CXCL8 (IL-8), CXCL9, CXCL10, CXCL11, CXCL12 Citation: Korbecki, J.; Kojder, K.; Kapczuk, P.; Kupnicka, P.; (SDF-1), CXCL13, CXCL14, CXCL15, CXCL16, CXCL17) as well as CXC chemokine receptors— Gawro´nska-Szklarz,B.; Gutowska, I.; CXCR1, CXCR2, CXCR3, CXCR4, CXCR5, CXCR6, CXCR7 and CXCR8. -

1 INVITED REVIEW Mechanisms of Gasdermin Family Members in Inflammasome Signaling and Cell Death Shouya Feng,* Daniel Fox,* Si M
INVITED REVIEW Mechanisms of Gasdermin family members in inflammasome signaling and cell death Shouya Feng,* Daniel Fox,* Si Ming Man Department of Immunology and Infectious Disease, The John Curtin School of Medical Research, The Australian National University, Canberra, Australia. * S.F. and D.F. equally contributed to this work Correspondence to Si Ming Man: Department of Immunology and Infectious Disease, The John Curtin School of Medical Research, The Australian National University, Canberra, 2601, Australia. [email protected] 1 Abstract The Gasdermin (GSDM) family consists of Gasdermin A (GSDMA), Gasdermin B (GSDMB), Gasdermin C (GSDMC), Gasdermin D (GSDMD), Gasdermin E (GSDME) and Pejvakin (PJVK). GSDMD is activated by inflammasome-associated inflammatory caspases. Cleavage of GSDMD by human or mouse caspase-1, human caspase-4, human caspase-5, and mouse caspase-11, liberates the N-terminal effector domain from the C-terminal inhibitory domain. The N-terminal domain oligomerizes in the cell membrane and forms a pore of 10-16 nm in diameter, through which substrates of a smaller diameter, such as interleukin (IL)-1β and IL- 18, are secreted. The increasing abundance of membrane pores ultimately leads to membrane rupture and pyroptosis, releasing the entire cellular content. Other than GSDMD, the N-terminal domain of all GSDMs, with the exception of PJVK, have the ability to form pores. There is evidence to suggest that GSDMB and GSDME are cleaved by apoptotic caspases. Here, we review the mechanistic functions of GSDM proteins with respect to their expression and signaling profile in the cell, with more focused discussions on inflammasome activation and cell death. -

Exploration of Prognostic Biomarkers and Therapeutic Targets in the Microenvironment of Bladder Cancer Based on CXC Chemokines
Exploration of Prognostic Biomarkers and Therapeutic Targets in The Microenvironment of Bladder Cancer Based on CXC Chemokines Xiaoqi Sun Department of Urology, Kaiping Central Hospital, Kaiping, 529300, China Qunxi Chen Department of Pathology, Sun Yat-sen University Cancer Center, Guangzhou, 510060, China Lihong Zhang Department of Pathology, Sun Yat-sen University Cancer Center, Guangzhou, 510060, China Jiewei Chen Department of Pathology, Sun Yat-sen University Cancer Center, Guangzhou, 510060, China Xinke Zhang ( [email protected] ) Sun Yat-sen University Cancer Center Research Keywords: Bladder cancer, Biomarkers, CXC Chemokines, Microenvironment Posted Date: February 24th, 2021 DOI: https://doi.org/10.21203/rs.3.rs-223127/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/29 Abstract Background: Bladder cancer (BLCA) has a high rate of morbidity and mortality, and is considered as one of the most malignant tumors of the urinary system. Tumor cells interact with surrounding interstitial cells, playing a key role in carcinogenesis and progression, which is partly mediated by chemokines. CXC chemokines exert anti‐tumor biological roles in the tumor microenvironment and affect patient prognosis. Nevertheless, their expression and prognostic values patients with BLCA remain unclear. Methods: We used online tools, including Oncomine, UALCAN, GEPIA, GEO databases, cBioPortal, GeneMANIA, DAVID 6.8, Metascape, TRUST (version 2.0), LinkedOmics, TCGA, and TIMER2.0 to perform the relevant analysis. Results: The mRNA levels of C-X-C motif chemokine ligand (CXCL)1, CXCL5, CXCL6, CXCL7, CXCL9, CXCL10, CXCL11, CXCL13, CXCL16, and CXCL17 were increased signicantly increased, and those of CXCL2, CXCL3, and CXCL12 were decreased signicantly in BLCA tissues as assessed using the Oncomine, TCGA, and GEO databases. -

Development and Validation of a Protein-Based Risk Score for Cardiovascular Outcomes Among Patients with Stable Coronary Heart Disease
Supplementary Online Content Ganz P, Heidecker B, Hveem K, et al. Development and validation of a protein-based risk score for cardiovascular outcomes among patients with stable coronary heart disease. JAMA. doi: 10.1001/jama.2016.5951 eTable 1. List of 1130 Proteins Measured by Somalogic’s Modified Aptamer-Based Proteomic Assay eTable 2. Coefficients for Weibull Recalibration Model Applied to 9-Protein Model eFigure 1. Median Protein Levels in Derivation and Validation Cohort eTable 3. Coefficients for the Recalibration Model Applied to Refit Framingham eFigure 2. Calibration Plots for the Refit Framingham Model eTable 4. List of 200 Proteins Associated With the Risk of MI, Stroke, Heart Failure, and Death eFigure 3. Hazard Ratios of Lasso Selected Proteins for Primary End Point of MI, Stroke, Heart Failure, and Death eFigure 4. 9-Protein Prognostic Model Hazard Ratios Adjusted for Framingham Variables eFigure 5. 9-Protein Risk Scores by Event Type This supplementary material has been provided by the authors to give readers additional information about their work. Downloaded From: https://jamanetwork.com/ on 10/02/2021 Supplemental Material Table of Contents 1 Study Design and Data Processing ......................................................................................................... 3 2 Table of 1130 Proteins Measured .......................................................................................................... 4 3 Variable Selection and Statistical Modeling ........................................................................................ -

Bio-Plex Pro™ Human Chemokine Panel
Acute Phase Response Cancer Cardiovascular Disease Cytokines, Chemokines, Growth Factors Diabetes Toxicology ™ Genotyping Bio-Plex Pro Immunoglobulin Isotyping Signal Transduction Human Chemokine Panel 6Ckine / CCL21, BCA-1 / CXCL13, CTACK / CCL27, ENA-78 / CXCL5, Eotaxin / CCL11, Eotaxin-2 / CCL24, Eotaxin-3 / CCL26, Fractalkine / CX3CL1, GCP-2 / CXCL6, GM-CSF, Gro-a / CXCL1, Gro-b / CXCL2, I-309 / CCL1, MAGNETIC SEPARATION ENABLED IFN-g, IL-1b, IL-2, IL-4, IL-6, IL-8 / CXCL8, IL-10, IL-16, IP-10 / CXCL10, I-TAC / CXCL11, MCP-1 / CCL2, MCP-2 / CCL8, MCP-3 / CCL7, MCP-4 / CCL13, MDC / CCL22, MIF, MIG / CXCL9, MIP-1a / CCL3, MIP-1d / CCL15, MIP-3a / CCL20, MIP-3b / CCL19, MPIF-1 / CCL23, SCYB16 / CXCL16, SDF-1a+b / CXCL12, TARC / CCL17, TECK / CCL25, TNF-a ■ All-in-one 40-plex kit High-Performance Multiplex Rigorous Assay Validation ■ Custom Immunoassays for Research All Bio-Plex Pro Assays undergo rigorous configurations The Bio-Plex Pro Human Chemokine evaluation that includes the following ■ Single level quality Panel features 40 magnetic bead–based parameters: control immunoassays to measure chemokines and ■■ Specificity (cross-reactivity) ■ Magnetic workflow chemotaxis-relevant analytes. The assays ■■ Accuracy (recovery) in key sample matrices have undergone rigorous evaluation to provide ■■ Inter- and intra-assay precision reliable performance for your research needs ■■ Sensitivity (limit of detection, LOD) and are available in flexible, customized formats. ■■ Assay working range (LLOQ/ULOQ) Chemokines are typically associated with the ■■ Linearity of dilution following areas of research: ■■ Parallelism and matrix effect ■■ Cancer/angiogenesis ■■ Performance characteristics in real samples ■■ Cardiovascular disease Assay Performance Definitions ■■ Autoimmune disorders The following parameters are indicative of ■■ Allergy/asthma assay performance, as shown in Table 1. -

The Metabolite α-KG Induces GSDMC-Dependent Pyroptosis Through Death Receptor 6-Activated Caspase-8
www.nature.com/cr www.cell-research.com ARTICLE OPEN The metabolite α-KG induces GSDMC-dependent pyroptosis through death receptor 6-activated caspase-8 Jia-yuan Zhang1, Bo Zhou1, Ru-yue Sun1, Yuan-li Ai1, Kang Cheng1, Fu-nan Li2, Bao-rui Wang2, Fan-jian Liu1, Zhi-hong Jiang1, Wei-jia Wang1, Dawang Zhou 1, Hang-zi Chen 1 and Qiao Wu 1 Pyroptosis is a form of regulated cell death mediated by gasdermin family members, among which the function of GSDMC has not been clearly described. Herein, we demonstrate that the metabolite α-ketoglutarate (α-KG) induces pyroptosis through caspase-8- mediated cleavage of GSDMC. Treatment with DM-αKG, a cell-permeable derivative of α-KG, elevates ROS levels, which leads to oxidation of the plasma membrane-localized death receptor DR6. Oxidation of DR6 triggers its endocytosis, and then recruits both pro-caspase-8 and GSDMC to a DR6 receptosome through protein-protein interactions. The DR6 receptosome herein provides a platform for the cleavage of GSDMC by active caspase-8, thereby leading to pyroptosis. Moreover, this α-KG-induced pyroptosis could inhibit tumor growth and metastasis in mouse models. Interestingly, the efficiency of α-KG in inducing pyroptosis relies on an acidic environment in which α-KG is reduced by MDH1 and converted to L-2HG that further boosts ROS levels. Treatment with lactic acid, the end product of glycolysis, builds an improved acidic environment to facilitate more production of L-2HG, which makes the originally pyroptosis-resistant cancer cells more susceptible to α-KG-induced pyroptosis. This study not only illustrates a pyroptotic pathway linked with metabolites but also identifies an unreported principal axis extending from ROS-initiated DR6 endocytosis to caspase-8-mediated cleavage of GSDMC for potential clinical application in tumor therapy. -

1 1 2 3 Genome-Wide Association Study of Body Fat Distribution
bioRxiv preprint doi: https://doi.org/10.1101/207498; this version posted July 13, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 2 3 4 Genome-Wide Association Study of Body Fat Distribution identifies Novel 5 Adiposity Loci and Sex-Specific Genetic Effects 6 7 Mathias Rask-Andersen*1, Torgny Karlsson1, Weronica E Ek1, Åsa Johansson*1 8 9 10 1Department of Immunology, Genetics and Pathology, Science for Life Laboratory, 11 Uppsala University. Box 256, 751 05, Uppsala, Sweden. 12 *Corresponding authors: [email protected] and 13 [email protected] 14 15 16 17 18 1 bioRxiv preprint doi: https://doi.org/10.1101/207498; this version posted July 13, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 19 Body mass and body fat composition are of clinical interest due to their links to 20 cardiovascular- and metabolic diseases. Fat stored in the trunk has been 21 suggested as more pathogenic compared to fat stored in other compartments of 22 the body. In this study, we performed genome-wide association studies (GWAS) 23 for the proportion of body fat distributed to the arms, legs and trunk estimated 24 from segmental bio-electrical impedance analysis (sBIA) for 362,499 individuals 25 from the UK Biobank. A total of 97 loci, were identified to be associated with 26 body fat distribution, 40 of which have not previously been associated with an 27 anthropometric trait. -

Cytokines Explored in Saliva and Tears from Radiated Cancer Patients Correlate with Clinical Manifestations, Influencing Importa
cells Article Cytokines Explored in Saliva and Tears from Radiated Cancer Patients Correlate with Clinical Manifestations, Influencing Important Immunoregulatory Cellular Pathways Lara A. Aqrawi 1,2 , Xiangjun Chen 1, Håvard Hynne 1, Cecilie Amdal 3, Sjur Reppe 4 , Hans Christian D. Aass 4, Morten Rykke 5, Lene Hystad Hove 5, Alix Young 5, Bente Brokstad Herlofson 1,6, Kristine Løken Westgaard 1,6, Tor Paaske Utheim 4,7,8,9, Hilde Kanli Galtung 8,* and Janicke Liaaen Jensen 1 1 Department of Oral Surgery and Oral Medicine, Faculty of Dentistry, University of Oslo, 0317 Oslo, Norway; [email protected] (L.A.A.); [email protected] (X.C.); [email protected] (H.H.); [email protected] (B.B.H.); [email protected] (K.L.W.); [email protected] (J.L.J.) 2 Department of Health Sciences, Kristiania University College, 0153 Oslo, Norway 3 Section for Head and Neck Oncology, Oslo University Hospital, 0379 Oslo, Norway; [email protected] 4 Department of Medical Biochemistry, Oslo University Hospital, 0450 Oslo, Norway; [email protected] (S.R.); [email protected] (H.C.D.A.); [email protected] (T.P.U.) 5 Department of Cariology and Gerodontology, Faculty of Dentistry, University of Oslo, 0455 Oslo, Norway; [email protected] (M.R.); [email protected] (L.H.H.); [email protected] (A.Y.) 6 Department of Otorhinolaryngology-Head and Neck Surgery Division for Head, Neck and Reconstructive Surgery, Oslo University Hospital, 0450 Oslo, Norway 7 Department of Plastic and Reconstructive -

And Nuclear Factor-Κb–Regulated CXC Chemokine Gene Expression in Lung Carcinogenesis
Cancer Prevention Research Cyclic AMP-Responsive Element Binding Protein– and Nuclear Factor-κB–Regulated CXC Chemokine Gene Expression in Lung Carcinogenesis Hongxia Sun, Wen-Cheng Chung, Seung-Hee Ryu, Zhenlin Ju, Hai T. Tran, Edward Kim, Jonathan M. Kurie and Ja Seok Koo Abstract The recognition of the importance of angiogenesis in tumor progression has led to the development of antiangiogenesis as a new strategy for cancer treatment and prevention. By modulating tumor microenvironment and inducing angiogenesis, the proinflammatory cytokine interleukine (IL)-1β has been reported to promote tumor development. However, the factors mediating IL-1β–induced angiogenesis in non–small cell lung cancer (NSCLC) and the regulation of these angiogenicfactorsby IL-1 β are less clear. Here, we report that IL-1β up-regulated an array of proangiogenic CXC chemokine genes in the NSCLC cell line A549 and in normal human tracheobronchial epithelium cells, as determined by microarray analysis. Further analysis revealed that IL-1β induced much higher protein levels of CXC chemokines in NSCLC cells than in normal human tracheobronchial epithelium cells. Con- ditioned medium from IL-1β–treated A549 cells markedly increased endothelial cell migra- tion, which was suppressed by neutralizing antibodies against CXCL5 and CXCR2. We also found that IL-1β–induced CXC chemokine gene overexpression in NSCLC cells was abro- gated with the knockdown of cyclic AMP-responsive element binding protein (CREB) or nuclear factor κB (NF-κB). Moreover, the expression of the CXC chemokine genes as well as CREB and NF-κB activities was greatly increased in the tumorigenic NSCLC cell line compared with normal, premalignant immortalized or nontumorigenic cell lines.