Exploration of Prognostic Biomarkers and Therapeutic Targets in the Microenvironment of Bladder Cancer Based on CXC Chemokines
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Prognostic Value of CXCL17 and CXCR8 Expression in Patients with Colon Cancer
ONCOLOGY LETTERS 20: 2711-2720, 2020 Prognostic value of CXCL17 and CXCR8 expression in patients with colon cancer HONGYAN YAO1,2, YUFENG LV3, XUEFENG BAI4, ZHAOJIN YU4 and XIAOJIAN LIU1 1Department of Pharmacology, School of Basic Medical Sciences, Jinzhou Medical University; 2Nuclear Medicine Department, Jinzhou Central Hospital, Jinzhou, Liaoning 121001; 3Department of Respiration and Critical Care, The Affiliated Hongqi Hospital of Mudanjiang Medical University, Mudanjiang, Heilongjiang 157011; 4Department of Pharmacology, School of Pharmacy, China Medical University, Shenyang, Liaoning 110001, P.R. China Received October 11, 2019; Accepted May 27, 2020 DOI: 10.3892/ol.2020.11819 Abstract. C-X-C motif chemokine ligand 17 (CXCL17) is a CXCL17/CXCR8 signalling may be involved in colon cancer mucous chemokine and its expression is highly correlated with and contribute to improved patient outcomes. that of G protein-coupled receptor 35 (GPR35), which has been confirmed as its receptor and named C‑X‑C motif chemokine Introduction receptor 8 (CXCR8). CXCL17 is upregulated in several types of cancer. However, the biological role of this chemokine Chemokines are a large family of cytokines serving important in colon cancer remains unknown. In the present study, the roles in the immune system, such as the regulation of cell traf- expression levels of CXCL17 and CXCR8 were examined ficking to inflammatory sites (1). By regulating the activity of using immunohistochemistry in 101 colon cancer tissues and cells in homeostasis and signal transduction, chemokines are 79 healthy tumour-adjacent tissues. CXCL17 and CXCR8 involved in numerous physiological and pathological processes expression levels were increased in the colon cancer samples such as angiogenesis, embryonic development, wound healing, compared with tumour-adjacent samples. -

Enhanced Monocyte Migration to CXCR3 and CCR5 Chemokines in COPD
ERJ Express. Published on March 10, 2016 as doi: 10.1183/13993003.01642-2015 ORIGINAL ARTICLE IN PRESS | CORRECTED PROOF Enhanced monocyte migration to CXCR3 and CCR5 chemokines in COPD Claudia Costa1, Suzanne L. Traves1, Susan J. Tudhope1, Peter S. Fenwick1, Kylie B.R. Belchamber1, Richard E.K. Russell2, Peter J. Barnes1 and Louise E. Donnelly1 Affiliations: 1Airway Disease, National Heart and Lung Institute, Imperial College London, London, UK. 2Chest Clinic, King Edward King VII Hospital, Windsor, UK. Correspondence: Louise E. Donnelly, Airway Disease, National Heart and Lung Institute, Dovehouse Street, London, SW3 6LY, UK. E-mail: [email protected] ABSTRACT Chronic obstructive pulmonary disease (COPD) patients exhibit chronic inflammation, both in the lung parenchyma and the airways, which is characterised by an increased infiltration of macrophages and T-lymphocytes, particularly CD8+ cells. Both cell types can express chemokine (C-X-C motif) receptor (CXCR)3 and C-C chemokine receptor 5 and the relevant chemokines for these receptors are elevated in COPD. The aim of this study was to compare chemotactic responses of lymphocytes and monocytes of nonsmokers, smokers and COPD patients towards CXCR3 ligands and chemokine (C-C motif) ligand (CCL)5. Migration of peripheral blood mononuclear cells, monocytes and lymphocytes from nonsmokers, smokers and COPD patients toward CXCR3 chemokines and CCL5 was analysed using chemotaxis assays. There was increased migration of peripheral blood mononuclear cells from COPD patients towards all chemokines studied when compared with nonsmokers and smokers. Both lymphocytes and monocytes contributed to this enhanced response, which was not explained by increased receptor expression. -

Cxcl17 and Its Association with T Cells
Hernández-Ruiz et al., Arch Autoimmune Archives of Autoimmune Diseases Dis 2020; 1(1):28-31. Commentary Cxcl17 and its association with T cells Marcela Hernández-Ruiz1, Albert Zlotnik2* 1Lymphotrek, San Diego, CA, United Keywords: CXCL17, Auto immunities, T cells, IL-23 States Abbreviations: BALF: Bronchoalveolar Lavage Fluid; CLP: Common Lymphoid Progenitor; CNS: 2Department of Physiology Central Nervous System; DP: Double Positive; EAE: Experimental Autoimmune Encephalomyelitis; LN: and Biophysics and Institute for Lymph Nodes; LTHSC: Long-term Hematopoietic Stem Cells; MHC: Major Histocompatibility Complex; Immunology, University of California Irvine, Irvine, CA, United States MOG: Myelin Oligodendrocyte Glycoprotein; STHCS: Short-Term Hematopoietic Stem Cells; TCR: T cell Receptor *Author for correspondence: Email: [email protected] We recently published an article describing the importance of CXCL17 in T cell responses [1]. In summary, we observed the following: Received date: September 02, 2020 Accepted date: September 23, 2020 1) Cxcl17 is necessary to maintain normal ratios of T cell subpopulations in lymph nodes (LN). 2) Cxcl17-/- mice develop more intense inflammatory responses than wild type mice. Copyright: © 2020 Hernández-Ruiz Cxcl17 is a mucosal chemokine predominantly expressed in mucosal tissues of the respiratory M, et al. This is an open-access article tract and digestive system [2-4]. However, it is also expressed in primary immune tissues such as distributed under the terms of the bone marrow and thymus (where its function is currently unknown- Table 1). Microarray data Creative Commons Attribution License, from the Immgen database of 298 purified immune cell subpopulations, and consistent with the which permits unrestricted use, distribution, and reproduction in any analysis shown in table 1, indicates that Cxcl17 is expressed in the thymus. -

Original Article Tocilizumab Infusion Therapy Normalizes Inflammation in Sporadic ALS Patients
Am J Neurodegener Dis 2013;2(2):129-139 www.AJND.us /ISSN:2165-591X/AJND1304002 Original Article Tocilizumab infusion therapy normalizes inflammation in sporadic ALS patients Milan Fiala1, Mathew T Mizwicki1, Rachel Weitzman1, Larry Magpantay2, Norihiro Nishimoto3 1Department of Surgery, David Geffen School of Medicine at UCLA, 100 UCLA Medical Plaza, Suite 220, Los Angeles, CA 90095-6970, USA; 2Department of Obstetrics and Gynecology, David Geffen School of Medicine at UCLA, Los Angeles, 650 Charles E. Young Drive, Los Angeles, CA, 90095-1735, USA; 3Department of Molecular Regulation for Intractable Diseases, Institute of Medical Sciences, Tokyo Medical University, Minamisenba, Chuo- ku, Osaka, 542-0081, Japan Received April 8 2013; Accepted May 19 2013; Epub June 21, 2013; Published July 1, 2013 Abstract: Patients with sporadic amyotrophic lateral sclerosis (sALS) show inflammation in the spinal cord and pe- ripheral blood. The inflammation is driven by stimulation of macrophages by aggregated superoxide dismutase 1 (SOD1) through caspase1, interleukin 1 (IL1), IL6 and chemokine signaling. Inflammatory gene activation is inhibit- ed in vitro by tocilizumab, a humanized antibody to IL6 receptor (IL6R). Tocilizumab inhibits global interleukin-6 (IL6) signaling, a key mechanism in chronic rheumatoid disorders. Here we studied in vivo baseline inflammatory gene transcription in peripheral blood mononuclear cells (PBMCs) of 10 sALS patients, and the effects of tocilizumab (ActemraR) infusions. At baseline, one half of ALS subjects had strong inflammatory activation (Group 1) (8 genes up regulated >4-fold, P<0.05 vs. controls) and the other half (Group 2) had weak activation. All patients showed greater than four-fold up regulation of MMP1, CCL7, CCL13 and CCL24. -
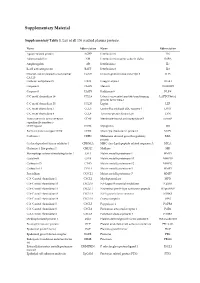
Supplementary Material
Supplementary Material Supplementary Table 1. List of all 136 studied plasma proteins. Name Abbreviation Name Abbreviation Agouti-related protein AGRP Interleukin-6 IL6 Adrenomedullin AM Interleukin-6 receptor subunit alpha IL6RA Amphiregulin AR Interleukin-7 IL7 B-cell activating factor BAFF Interleukin-8 IL8 Ovarian cancer-related tumor marker CA125 Immunoglobulin-like transcript 3 ILT3 CA 125 Carbonic anhydrase IX CAIX Integrin alpha-1 ITGA1 Caspase-3 CASP3 Melusin ITGB1BP2 Caspase-8 CASP8 Kallikrein-6 KLK6 C-C motif chemokine 19 CCL19 Latency-associated peptide transforming LAPTGFbeta1 growth factor beta-1 C-C motif chemokine 20 CCL20 Leptin LEP C-C motif chemokine 3 CCL3 Lectin-like oxidized LDL receptor 1 LOX1 C-C motif chemokine 4 CCL4 Tyrosine-protein kinase Lyn LYN Tumor necrosis factor receptor CD40 Membrane-bound aminopeptidase P mAmP superfamily member 5 CD40 ligand CD40L Myoglobin MB Early activation antigen CD69 CD69 Monocyte chemotactic protein 1 MCP1 Cadherin-3 CDH3 Melanoma-derived growth regulatory MIA protein Cyclin-dependent kinase inhibitor 1 CDKN1A MHC class I polypeptide-related sequence A MICA Chitinase-3-like protein 1 CHI3L1 Midkine MK Macrophage colony-stimulating factor 1 CSF1 Matrix metalloproteinase-1 MMP1 Cystatin-B CSTB Matrix metalloproteinase-10 MMP10 Cathepsin D CTSD Matrix metalloproteinase-12 MMP12 Cathepsin L1 CTSL1 Matrix metalloproteinase-3 MMP3 Fractalkine CX3CL1 Matrix metalloproteinase-7 MMP7 C-X-C motif chemokine 1 CXCL1 Myeloperoxidase MPO C-X-C motif chemokine 10 CXCL10 NF-kappa-B essential -

The CXCL7/CXCR1/2 Axis Is a Key Driver in the Growth of Clear Cell Renal Cell Carcinoma
Published OnlineFirst December 12, 2013; DOI: 10.1158/0008-5472.CAN-13-1267 Cancer Therapeutics, Targets, and Chemical Biology Research The CXCL7/CXCR1/2 Axis Is a Key Driver in the Growth of Clear Cell Renal Cell Carcinoma Renaud Grepin 5,Melanie Guyot1, Sandy Giuliano1, Marina Boncompagni1, Damien Ambrosetti1,2, Emmanuel Chamorey3, Jean-Yves Scoazec4, Sylvie Negrier4,Hel ene Simonnet4, and Gilles Pages 1 Abstract Mutations in the von Hippel–Lindau gene upregulate expression of the central angiogenic factor VEGF, which drives abnormal angiogenesis in clear cell renal cell carcinomas (ccRCC). However, the overexpression of VEGF in these tumors was not found to correlate with overall survival. Here, we show that the proangiogenic, proin- flammatory cytokine CXCL7 is an independent prognostic factor for overall survival in this setting. CXCL7 antibodies strongly reduced the growth of ccRCC tumors in nude mice. Conversely, conditional overexpression of CXCL7 accelerated ccRCC development. CXCL7 promoted cell proliferation in vivo and in vitro, in which expression of CXCL7 was induced by the central proinflammatory cytokine interleukin (IL)-1b. ccRCC cells normally secrete low amounts of CXCL7; it was more highly expressed in tumors due to high levels of IL-1b there. We found that a pharmacological inhibitor of the CXCL7 receptors CXCR1 and CXCR2 (SB225002) was sufficient to inhibit endothelial cell proliferation and ccRCC growth. Because CXCR1 and CXCR2 are present on both endothelial and ccRCC cells, their inhibition affected both the tumor vasculature and the proliferation of tumor cells. Our results highlight the CXCL7/CXCR1/CXCR2 axis as a pertinent target for the treatment of ccRCC. -

Mice in Inflammation Psoriasiform Skin IL-22 Is Required for Imiquimod-Induced
IL-22 Is Required for Imiquimod-Induced Psoriasiform Skin Inflammation in Mice Astrid B. Van Belle, Magali de Heusch, Muriel M. Lemaire, Emilie Hendrickx, Guy Warnier, Kyri This information is current as Dunussi-Joannopoulos, Lynette A. Fouser, Jean-Christophe of September 29, 2021. Renauld and Laure Dumoutier J Immunol published online 30 November 2011 http://www.jimmunol.org/content/early/2011/11/30/jimmun ol.1102224 Downloaded from Why The JI? Submit online. http://www.jimmunol.org/ • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average by guest on September 29, 2021 Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2011 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. Published November 30, 2011, doi:10.4049/jimmunol.1102224 The Journal of Immunology IL-22 Is Required for Imiquimod-Induced Psoriasiform Skin Inflammation in Mice Astrid B. Van Belle,*,† Magali de Heusch,*,† Muriel M. Lemaire,*,† Emilie Hendrickx,*,† Guy Warnier,*,† Kyri Dunussi-Joannopoulos,‡ Lynette A. Fouser,‡ Jean-Christophe Renauld,*,†,1 and Laure Dumoutier*,†,1 Psoriasis is a common chronic autoimmune skin disease of unknown cause that involves dysregulated interplay between immune cells and keratinocytes. -

Induced CNS Expression of CXCL1 Augments Neurologic Disease in a Murine Model of Multiple Sclerosis Via Enhanced Neutrophil Recruitment
Eur. J. Immunol. 2018. 48: 1199–1210 DOI: 10.1002/eji.201747442 Jonathan J. Grist et al. 1199 Basic Immunodeficiencies and autoimmunity Research Article Induced CNS expression of CXCL1 augments neurologic disease in a murine model of multiple sclerosis via enhanced neutrophil recruitment Jonathan J. Grist1, Brett S. Marro3, Dominic D. Skinner1, Amber R. Syage1, Colleen Worne1, Daniel J. Doty1, Robert S. Fujinami1,2 andThomasE.Lane1,2 1 Department of Pathology, Division of Microbiology and Immunology, University of Utah, School of Medicine, Salt Lake City, UT, USA 2 Immunology, Inflammation, and Infectious Disease Initiative, University of Utah, UT, USA 3 Department of Molecular Biology and Biochemistry, University of California, Irvine, CA, USA Increasing evidence points to an important role for neutrophils in participating in the pathogenesis of the human demyelinating disease MS and the animal model EAE. There- fore, a better understanding of the signals controlling migration of neutrophils as well as evaluating the role of these cells in demyelination is important to define cellular com- ponents that contribute to disease in MS patients. In this study, we examined the func- tional role of the chemokine CXCL1 in contributing to neuroinflammation and demyeli- nation in EAE. Using transgenic mice in which expression of CXCL1 is under the control of a tetracycline-inducible promoter active within glial fibrillary acidic protein-positive cells, we have shown that sustained CXCL1 expression within the CNS increased the severity of clinical and histologic disease that was independent of an increase in the frequency of encephalitogenic Th1 and Th17 cells. Rather, disease was associated with enhanced recruitment of CD11b+Ly6G+ neutrophils into the spinal cord. -

The Effect of Hypoxia on the Expression of CXC Chemokines and CXC Chemokine Receptors—A Review of Literature
International Journal of Molecular Sciences Review The Effect of Hypoxia on the Expression of CXC Chemokines and CXC Chemokine Receptors—A Review of Literature Jan Korbecki 1 , Klaudyna Kojder 2, Patrycja Kapczuk 1, Patrycja Kupnicka 1 , Barbara Gawro ´nska-Szklarz 3 , Izabela Gutowska 4 , Dariusz Chlubek 1 and Irena Baranowska-Bosiacka 1,* 1 Department of Biochemistry and Medical Chemistry, Pomeranian Medical University in Szczecin, Powsta´nców Wielkopolskich 72 Av., 70-111 Szczecin, Poland; [email protected] (J.K.); [email protected] (P.K.); [email protected] (P.K.); [email protected] (D.C.) 2 Department of Anaesthesiology and Intensive Care, Pomeranian Medical University in Szczecin, Unii Lubelskiej 1, 71-281 Szczecin, Poland; [email protected] 3 Department of Pharmacokinetics and Therapeutic Drug Monitoring, Pomeranian Medical University in Szczecin, Powsta´nców Wielkopolskich 72 Av., 70-111 Szczecin, Poland; [email protected] 4 Department of Medical Chemistry, Pomeranian Medical University in Szczecin, Powsta´nców Wlkp. 72 Av., 70-111 Szczecin, Poland; [email protected] * Correspondence: [email protected]; Tel.: +48-914661515 Abstract: Hypoxia is an integral component of the tumor microenvironment. Either as chronic or cycling hypoxia, it exerts a similar effect on cancer processes by activating hypoxia-inducible factor-1 (HIF-1) and nuclear factor (NF-κB), with cycling hypoxia showing a stronger proinflammatory influ- ence. One of the systems affected by hypoxia is the CXC chemokine system. This paper reviews all available information on hypoxia-induced changes in the expression of all CXC chemokines (CXCL1, CXCL2, CXCL3, CXCL4, CXCL5, CXCL6, CXCL7, CXCL8 (IL-8), CXCL9, CXCL10, CXCL11, CXCL12 Citation: Korbecki, J.; Kojder, K.; Kapczuk, P.; Kupnicka, P.; (SDF-1), CXCL13, CXCL14, CXCL15, CXCL16, CXCL17) as well as CXC chemokine receptors— Gawro´nska-Szklarz,B.; Gutowska, I.; CXCR1, CXCR2, CXCR3, CXCR4, CXCR5, CXCR6, CXCR7 and CXCR8. -

IL-34–Dependent Intrarenal and Systemic Mechanisms Promote Lupus Nephritis in MRL-Faslpr Mice
BASIC RESEARCH www.jasn.org IL-34–Dependent Intrarenal and Systemic Mechanisms Promote Lupus Nephritis in MRL-Faslpr Mice Yukihiro Wada,1 Hilda M. Gonzalez-Sanchez,1 Julia Weinmann-Menke,2 Yasunori Iwata,1 Amrendra K. Ajay,1 Myriam Meineck,2 and Vicki R. Kelley1 1Renal Division, Department of Medicine, Brigham and Women’s Hospital, Boston, Massachusetts; and 2Department of Nephrology and Rheumatology, University Medical Center of the Johannes Gutenberg University Mainz, Mainz, Germany ABSTRACT lpr Background In people with SLE and in the MRL-Fas lupus mouse model, macrophages and autoanti- bodies are central to lupus nephritis. IL-34 mediates macrophage survival and proliferation, is expressed by tubular epithelial cells (TECs), and binds to the cFMS receptor on macrophages and to a newly identified second receptor, PTPRZ. Methods To investigate whether IL-34–dependent intrarenal and systemic mechanisms promote lupus lpr nephritis, we compared lupus nephritis and systemic illness in MRL-Fas mice expressing IL-34 and IL-34 lpr knockout (KO) MRL-Fas mice. We also assessed expression of IL-34 and the cFMS and PTPRZ receptors in patients with lupus nephritis. lpr Results Intrarenal IL-34 and its two receptors increase during lupus nephritis in MRL-Fas mice. In knock- out mice lacking IL-34, nephritis and systemic illness are suppressed. IL-34 fosters intrarenal macrophage accumulation via monocyte proliferation in bone marrow (which increases circulating monocytes that are recruited by chemokines into the kidney) and via intrarenal macrophage proliferation. This accumulation leads to macrophage-mediated TEC apoptosis. We also found suppression of circulating autoantibodies and glomerular antibody deposits in the knockout mice. -

The Chemokine System in Innate Immunity
Downloaded from http://cshperspectives.cshlp.org/ on September 28, 2021 - Published by Cold Spring Harbor Laboratory Press The Chemokine System in Innate Immunity Caroline L. Sokol and Andrew D. Luster Center for Immunology & Inflammatory Diseases, Division of Rheumatology, Allergy and Immunology, Massachusetts General Hospital, Harvard Medical School, Boston, Massachusetts 02114 Correspondence: [email protected] Chemokines are chemotactic cytokines that control the migration and positioning of immune cells in tissues and are critical for the function of the innate immune system. Chemokines control the release of innate immune cells from the bone marrow during homeostasis as well as in response to infection and inflammation. Theyalso recruit innate immune effectors out of the circulation and into the tissue where, in collaboration with other chemoattractants, they guide these cells to the very sites of tissue injury. Chemokine function is also critical for the positioning of innate immune sentinels in peripheral tissue and then, following innate immune activation, guiding these activated cells to the draining lymph node to initiate and imprint an adaptive immune response. In this review, we will highlight recent advances in understanding how chemokine function regulates the movement and positioning of innate immune cells at homeostasis and in response to acute inflammation, and then we will review how chemokine-mediated innate immune cell trafficking plays an essential role in linking the innate and adaptive immune responses. hemokines are chemotactic cytokines that with emphasis placed on its role in the innate Ccontrol cell migration and cell positioning immune system. throughout development, homeostasis, and in- flammation. The immune system, which is de- pendent on the coordinated migration of cells, CHEMOKINES AND CHEMOKINE RECEPTORS is particularly dependent on chemokines for its function. -

Starvation and Antimetabolic Therapy Promote Cytokine Release and Recruitment of Immune Cells
Starvation and antimetabolic therapy promote cytokine release and recruitment of immune cells Franziska Püschela, Francesca Favaroa,b,c,d,1, Jaime Redondo-Pedrazaa,1, Estefanía Lucendoa, Raffaella Iurlaroa, Sandrine Marchettie, Blanca Majema, Eric Elderingb,c,d, Ernest Nadalf, Jean-Ehrland Riccie, Eric Chevetg,h, and Cristina Muñoz-Pinedoa,i,2 aOncobell Program, Bellvitge Biomedical Research Institute, Hospitalet, 08908 Barcelona, Spain; bDepartment of Experimental Immunology, Amsterdam University Medical Centers, University of Amsterdam, 1105 AZ Amsterdam, The Netherlands; cLymphoma and Myeloma Center, Cancer Center Amsterdam, University of Amsterdam, 1105 AZ Amsterdam, The Netherlands; dAmsterdam Institute for Infection & Immunity, 1105 AZ Amsterdam, The Netherlands; eINSERM, Centre Méditerranéen de Médecine Moléculaire, Université Côte d’Azur, 06204 Nice, France; fThoracic Oncology Unit, Department of Medical Oncology, Catalan Institute of Oncology, Hospitalet, 08908 Barcelona, Spain; gINSERM U1242 “Chemistry, Oncogenesis, Stress, Signaling,” Université de Rennes, 35042 Rennes, France; hINSERM U1242, Centre de Lutte Contre le Cancer Eugène Marquis, 35042 Rennes, France; and iDepartment of Basic Nursing, Faculty of Medicine and Health Sciences, Universitat de Barcelona, Hospitalet, 08907 Barcelona, Spain Edited by Karen H. Vousden, Francis Crick Institute, London, United Kingdom, and approved March 16, 2020 (received for review August 14, 2019) Cellular starvation is typically a consequence of tissue injury that of oxygen or nutrients. However, some reports suggest that nutrient disrupts the local blood supply but can also occur where cell restriction, even without cell death, can be sufficient to promote the populations outgrow the local vasculature, as observed in solid synthesis and/or secretion of select proinflammatory cytokines tumors. Cells react to nutrient deprivation by adapting their (7–9).