University of California, San Diego
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Arctic Picoeukaryote Micromonas Pusilla Benefits
Biogeosciences Discuss., https://doi.org/10.5194/bg-2018-28 Manuscript under review for journal Biogeosciences Discussion started: 5 February 2018 c Author(s) 2018. CC BY 4.0 License. 1 The Arctic picoeukaryote Micromonas pusilla benefits 2 synergistically from warming and ocean acidification 3 4 Clara J. M. Hoppe1,2*, Clara M. Flintrop1,3 and Björn Rost1 5 6 1 Marine Biogeosciences, Alfred Wegener Institute – Helmholtz Centre for Polar and Marine 7 Research, 27570 Bremerhaven, Germany 8 2 Norwegian Polar Institute, 9296 Tromsø, Norway 9 3 MARUM, 28359 Bremen, Germany 10 11 *Correspondence to: Clara J. M. Hoppe ([email protected] 12 13 14 15 Abstract 16 In the Arctic Ocean, climate change effects such as warming and ocean acidification (OA) are 17 manifesting faster than in other regions. Yet, we are lacking a mechanistic understanding of the 18 interactive effects of these drivers on Arctic primary producers. In the current study, one of the 19 most abundant species of the Arctic Ocean, the prasinophyte Micromonas pusilla, was exposed 20 to a range of different pCO2 levels at two temperatures representing realistic scenarios for 21 current and future conditions. We observed that warming and OA synergistically increased 22 growth rates at intermediate to high pCO2 levels. Furthermore, elevated temperatures shifted 23 the pCO2-optimum of biomass production to higher levels. Based on changes in cellular 24 composition and photophysiology, we hypothesise that the observed synergies can be explained 25 by beneficial effects of warming on carbon fixation in combination with facilitated carbon 26 acquisition under OA. Our findings help to understand the higher abundances of picoeukaryotes 27 such as M. -

ABSTRACT Gregarine Parasitism in Dragonfly Populations of Central
ABSTRACT Gregarine Parasitism in Dragonfly Populations of Central Texas with an Assessment of Fitness Costs in Erythemis simplicicollis Jason L. Locklin, Ph.D. Mentor: Darrell S. Vodopich, Ph.D. Dragonfly parasites are widespread and frequently include gregarines (Phylum Apicomplexa) in the gut of the host. Gregarines are ubiquitous protozoan parasites that infect arthropods worldwide. More than 1,600 gregarine species have been described, but only a small percentage of invertebrates have been surveyed for these apicomplexan parasites. Some consider gregarines rather harmless, but recent studies suggest otherwise. Odonate-gregarine studies have more commonly involved damselflies, and some have considered gregarines to rarely infect dragonflies. In this study, dragonfly populations were surveyed for gregarines and an assessment of fitness costs was made in a common and widespread host species, Erythemis simplicicollis. Adult dragonfly populations were surveyed weekly at two reservoirs in close proximity to one another and at a flow-through wetland system. Gregarine prevalences and intensities were compared within host populations between genders, among locations, among wing loads, and through time. Host fitness parameters measured included wing load, egg size, clutch size, and total egg count. Of the 37 dragonfly species surveyed, 14 species (38%) hosted gregarines. Thirteen of those species were previously unreported as hosts. Gregarine prevalences ranged from 2% – 52%. Intensities ranged from 1 – 201. Parasites were aggregated among their hosts. Gregarines were found only in individuals exceeding a minimum wing load, indicating that gregarines are likely not transferred from the naiad to adult during emergence. Prevalence and intensity exhibited strong seasonality during both years at one of the reservoirs, but no seasonal trend was detected at the wetland. -
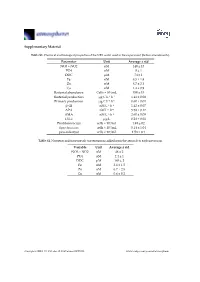
Supplementary Material Parameter Unit Average ± Std NO3 + NO2 Nm
Supplementary Material Table S1. Chemical and biological properties of the NRS water used in the experiment (before amendments). Parameter Unit Average ± std NO3 + NO2 nM 140 ± 13 PO4 nM 8 ± 1 DOC μM 74 ± 1 Fe nM 8.5 ± 1.8 Zn nM 8.7 ± 2.1 Cu nM 1.4 ± 0.9 Bacterial abundance Cells × 104/mL 350 ± 15 Bacterial production μg C L−1 h−1 1.41 ± 0.08 Primary production μg C L−1 h−1 0.60 ± 0.01 β-Gl nM L−1 h−1 1.42 ± 0.07 APA nM L−1 h−1 5.58 ± 0.17 AMA nM L−1·h−1 2.60 ± 0.09 Chl-a μg/L 0.28 ± 0.01 Prochlorococcus cells × 104/mL 1.49 ± 02 Synechococcus cells × 104/mL 5.14 ± 1.04 pico-eukaryot cells × 103/mL 1.58 × 0.1 Table S2. Nutrients and trace metals concentrations added from the aerosols to each mesocosm. Variable Unit Average ± std NO3 + NO2 nM 48 ± 2 PO4 nM 2.4 ± 1 DOC μM 165 ± 2 Fe nM 2.6 ± 1.5 Zn nM 6.7 ± 2.5 Cu nM 0.6 ± 0.2 Atmosphere 2019, 10, 358; doi:10.3390/atmos10070358 www.mdpi.com/journal/atmosphere Atmosphere 2019, 10, 358 2 of 6 Table S3. ANOVA test results between control, ‘UV-treated’ and ‘live-dust’ treatments at 20 h or 44 h, with significantly different values shown in bold. ANOVA df Sum Sq Mean Sq F Value p-value Chl-a 20 H 2, 6 0.03, 0.02 0.02, 0 4.52 0.0634 44 H 2, 6 0.02, 0 0.01, 0 23.13 0.002 Synechococcus Abundance 20 H 2, 7 8.23 × 107, 4.11 × 107 4.11 × 107, 4.51 × 107 0.91 0.4509 44 H 2, 7 5.31 × 108, 6.97 × 107 2.65 × 108, 1.16 × 107 22.84 0.0016 Prochlorococcus Abundance 20 H 2, 8 4.22 × 107, 2.11 × 107 2.11 × 107, 2.71 × 106 7.77 0.0216 44 H 2, 8 9.02 × 107, 1.47 × 107 4.51 × 107, 2.45 × 106 18.38 0.0028 Pico-eukaryote -

2018 Strassert JFH, Hehenberger E, Del Campo J, Okamoto N, Kolisko M
2018 Strassert JFH, Hehenberger E, del Campo J, Okamoto N, Kolisko M, Richards TA, Worden AZ, Santoro AE & PJ Keeling. Phylogeny, evidence for a cryptic plastid, and distribution of Chytriodinium parasites (Dinophyceae) infecting copepods. Journal of Eukaryotic Microbiology. https://doi.org/10.1111/jeu.12701 Joo S, Wang MH, Lui G, Lee J, Barnas A, Kim E, Sudek S, Worden AZ & JH Lee. Common ancestry of heterodimerizing TALE homeobox transcription factors across Metazoa and Archaeplastida. BMC Biology. 16:136. doi: 10.1186/s12915-018-0605-5 Bachy C, Charlesworth CJ, Chan AM, Finke JF, Wong C-H, Wei C-L, Sudek S, Coleman ML, Suttle CA & AZ Worden. Transcriptional responses of the marine green alga Micromonas pusilla and an infecting prasinovirus under different phosphate conditions. Environmental Microbiology. Vol 20:2898-2912. Guo J, Wilken S, Jimenez V, Choi CJ, Ansong CK, Dannebaum R, Sudek L, Milner D, Bachy C, Reistetter EN, Elrod VA, Klimov D, Purvine SO, Wei C-L, Kunde-Ramamoorthy G, Richards TA, Goodenough U, Smith RD, Callister SJ & AZ Worden. Specialized proteomic responses and an ancient photoprotection mechanism sustain marine green algal growth during phosphate limitation. Nature Microbiology. Vol 3:781–790. Okamoto N, Gawryluk RMR, del Campo J, Strassert JFH, Lukeš J, Richards TA, Worden AZ, Santoro AE & PJ Keeling. A revised taxonomy of diplonemids Including the Eupelagonemidae n. fam. and a Type Species, Eupelagonema oceanica n. gen. & sp. The Journal of Eukaryotic Microbiology. https://doi.org/10.1111/jeu.12679 Orsi WD, Wilken S, del Campo J, Heger T, James E, Richards TA, Keeling PJ, Worden AZ & AE. -

Spatial Variability of Picoeukaryotic Communities in the Mariana Trench Hongmei Jing1, Yue Zhang1,2, Yingdong Li3, Wenda Zhu1,2 & Hongbin Liu 3
www.nature.com/scientificreports OPEN Spatial Variability of Picoeukaryotic Communities in the Mariana Trench Hongmei Jing1, Yue Zhang1,2, Yingdong Li3, Wenda Zhu1,2 & Hongbin Liu 3 Picoeukaryotes play prominent roles in the biogeochemical cycles in marine ecosystems. However, their Received: 14 June 2018 molecular diversity studies have been confned in marine surface waters or shallow coastal sediments. Accepted: 5 October 2018 Here, we investigated the diversity and metabolic activity of picoeukaryotic communities at depths Published: xx xx xxxx ranging from the surface to the abyssopelagic zone in the western Pacifc Ocean above the north and south slopes of the Mariana Trench. This was achieved by amplifying and sequencing the V4 region of both 18S ribosomal DNA and cDNA using Illumina HiSeq sequencing. Our study revealed: (1) Four super-groups (i.e., Alveolata, Opisthokonta, Rhizaria and Stramenopiles) dominated the picoeukaryote assemblages through the water column, although they accounted for diferent proportions at DNA and cDNA levels. Our data expand the deep-sea assemblages from current bathypelagic to abyssopelagic zones. (2) Using the cDNA-DNA ratio as a proxy of relative metabolic activity, the highest activity for most subgroups was usually found in the mesopelagic zone; and (3) Population shift along the vertical scale was more prominent than that on the horizontal diferences, which might be explained by the sharp physicochemical gradients along the water depths. Overall, our study provides a better understanding of the diversity and metabolic activity of picoeukaryotes in water columns of the deep ocean in response to varying environmental conditions. Marine picoeukaryotes, (i.e., picoplanktonic eukaryotes of <2 μm in size), are capable of photosynthetic, hetero- trophic and mixotrophic metabolisms1. -

Supplementary Figure 1 Multicenter Randomised Control Trial 2746 Randomised
Supplementary Figure 1 Multicenter randomised control trial 2746 randomised 947 control 910 MNP without zinc 889 MNP with zinc 223 lost to follow up 219 lost to follow up 183 lost to follow up 34 refused 29 refused 37 refused 16 died 12 died 9 died 3 excluded 4 excluded 1 excluded 671 in follow-up 646 in follow-up 659 in follow-up at 24mo of age at 24mo of age at 24mo of age Selection for Microbiome sequencing 516 paired samples unavailable 469 paired samples unavailable 497 paired samples unavailable 69 antibiotic use 63 antibiotic use 67 antibiotic use 31 outside of WLZ criteria 37 outside of WLZ criteria 34 outside of WLZ criteria 6 diarrhea last 7 days 2 diarrhea last 7 days 7 diarrhea last 7 days 39 WLZ > -1 at 24 mo 10 WLZ < -2 at 24mo 58 WLZ > -1 at 24 mo 17 WLZ < -2 at 24mo 48 WLZ > -1 at 24 mo 8 WLZ < -2 at 24mo available for selection available for selection available for selection available for selection available for selection1 available for selection1 14 selected 10 selected 15 selected 14 selected 20 selected1 7 selected1 1 Two subjects (one in the reference WLZ group and one undernourished) had, at 12 months, no diarrhea within 1 day of stool collection but reported diarrhea within 7 days prior. Length, cm kg Weight, Supplementary Figure 2. Length (left) and weight (right) z-scores of children recruited into clinical trial NCT00705445 during the first 24 months of life. Median and quantile values are shown, with medians for participants profiled in current study indicated by red (undernourished) and black (reference WLZ) lines. -

Why the –Omic Future of Apicomplexa Should Include Gregarines Julie Boisard, Isabelle Florent
Why the –omic future of Apicomplexa should include Gregarines Julie Boisard, Isabelle Florent To cite this version: Julie Boisard, Isabelle Florent. Why the –omic future of Apicomplexa should include Gregarines. Biology of the Cell, Wiley, 2020, 10.1111/boc.202000006. hal-02553206 HAL Id: hal-02553206 https://hal.archives-ouvertes.fr/hal-02553206 Submitted on 24 Apr 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Article title: Why the –omic future of Apicomplexa should include Gregarines. Names of authors: Julie BOISARD1,2 and Isabelle FLORENT1 Authors affiliations: 1. Molécules de Communication et Adaptation des Microorganismes (MCAM, UMR 7245), Département Adaptations du Vivant (AVIV), Muséum National d’Histoire Naturelle, CNRS, CP52, 57 rue Cuvier 75231 Paris Cedex 05, France. 2. Structure et instabilité des génomes (STRING UMR 7196 CNRS / INSERM U1154), Département Adaptations du vivant (AVIV), Muséum National d'Histoire Naturelle, CP 26, 57 rue Cuvier 75231 Paris Cedex 05, France. Short Title: Gregarines –omics for Apicomplexa studies -

Protistology an International Journal Vol
Protistology An International Journal Vol. 10, Number 2, 2016 ___________________________________________________________________________________ CONTENTS INTERNATIONAL SCIENTIFIC FORUM «PROTIST–2016» Yuri Mazei (Vice-Chairman) Welcome Address 2 Organizing Committee 3 Organizers and Sponsors 4 Abstracts 5 Author Index 94 Forum “PROTIST-2016” June 6–10, 2016 Moscow, Russia Website: http://onlinereg.ru/protist-2016 WELCOME ADDRESS Dear colleagues! Republic) entitled “Diplonemids – new kids on the block”. The third lecture will be given by Alexey The Forum “PROTIST–2016” aims at gathering Smirnov (Saint Petersburg State University, Russia): the researchers in all protistological fields, from “Phylogeny, diversity, and evolution of Amoebozoa: molecular biology to ecology, to stimulate cross- new findings and new problems”. Then Sandra disciplinary interactions and establish long-term Baldauf (Uppsala University, Sweden) will make a international scientific cooperation. The conference plenary presentation “The search for the eukaryote will cover a wide range of fundamental and applied root, now you see it now you don’t”, and the fifth topics in Protistology, with the major focus on plenary lecture “Protist-based methods for assessing evolution and phylogeny, taxonomy, systematics and marine water quality” will be made by Alan Warren DNA barcoding, genomics and molecular biology, (Natural History Museum, United Kingdom). cell biology, organismal biology, parasitology, diversity and biogeography, ecology of soil and There will be two symposia sponsored by ISoP: aquatic protists, bioindicators and palaeoecology. “Integrative co-evolution between mitochondria and their hosts” organized by Sergio A. Muñoz- The Forum is organized jointly by the International Gómez, Claudio H. Slamovits, and Andrew J. Society of Protistologists (ISoP), International Roger, and “Protists of Marine Sediments” orga- Society for Evolutionary Protistology (ISEP), nized by Jun Gong and Virginia Edgcomb. -

High Abundance of Salps in the Coastal Gulf of Alaska During 2011
See discussions, stats, and author profiles for this publication at: https://www.researchgate.net/publication/301733881 High abundance of salps in the coastal Gulf of Alaska during 2011: A first record of bloom occurrence for the northern Gulf Article in Deep Sea Research Part II Topical Studies in Oceanography · April 2016 DOI: 10.1016/j.dsr2.2016.04.009 CITATION READS 1 33 4 authors, including: Ayla J. Doubleday Moira Galbraith University of Alaska System Institute of Ocean Sciences, Sidney, BC, Cana… 3 PUBLICATIONS 16 CITATIONS 28 PUBLICATIONS 670 CITATIONS SEE PROFILE SEE PROFILE Some of the authors of this publication are also working on these related projects: l. salmonis and c. clemensi attachment sites: chum salmon View project All content following this page was uploaded by Moira Galbraith on 10 June 2016. The user has requested enhancement of the downloaded file. All in-text references underlined in blue are added to the original document and are linked to publications on ResearchGate, letting you access and read them immediately. Deep-Sea Research II ∎ (∎∎∎∎) ∎∎∎–∎∎∎ Contents lists available at ScienceDirect Deep-Sea Research II journal homepage: www.elsevier.com/locate/dsr2 High abundance of salps in the coastal Gulf of Alaska during 2011: A first record of bloom occurrence for the northern Gulf Kaizhi Li a, Ayla J. Doubleday b, Moira D. Galbraith c, Russell R. Hopcroft b,n a Key Laboratory of Tropical Marine Bio-resources and Ecology, South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou 510301, China b Institute of Marine Science, University of Alaska, Fairbanks, AK 99775-7220, USA c Institute of Ocean Sciences, Fisheries and Oceans Canada, P.O. -

Assessing the Efficiency of Molecular Markers for the Species Identification of Gregarines Isolated from the Mealworm and Super Worm Midgut
Microorganisms 2018, 6, 119 1 of 20 Article Assessing the Efficiency of Molecular Markers for the Species Identification of Gregarines Isolated from the Mealworm and Super Worm Midgut Chiara Nocciolini 1, Claudio Cucini 2, Chiara Leo 2, Valeria Francardi 3, Elena Dreassi 4 and Antonio Carapelli 2,* 1 Polo d’Innovazione di Genomica Genetica e Biologia, 53100 Siena, Italy; [email protected] 2 Department of Life Sciences, University of Siena, 53100 Siena, Italy; [email protected] (C.C.); [email protected] (C.L.) 3 Consiglio per la ricerca in agricoltura e analisi dell’economia agraria – Centro di Difesa e Certificazione CREA-DC) Research Centre for Plant Protection and Certification, via di Lanciola 12/A, 50125 Firenze, Italy; [email protected] 4 Department of Biotechnology, Chemistry and Pharmacy, University of Siena, 53100 Siena, Italy; [email protected] * Correspondence: [email protected]; Tel.: +39-0577-234-410 Received: 28 September 2018; Accepted: 23 November 2018; Published: 27 November 2018 Abstract: Protozoa, of the taxon Gregarinasina, are a heterogeneous group of Apicomplexa that includes ~1600 species. They are parasites of a large variety of both marine and terrestrial invertebrates, mainly annelids, arthropods and mollusks. Unlike coccidians and heamosporidians, gregarines have not proven to have a negative effect on human welfare; thus, they have been poorly investigated. This study focuses on the molecular identification and phylogeny of the gregarine species found in the midgut of two insect species that are considered as an alternative source of animal proteins for the human diet: the mealworm Tenebrio molitor, and the super-worm Zophobas atratus (Coleoptera: Tenebrionidae). -

Rhythmicity of Coastal Marine Picoeukaryotes, Bacteria and Archaea Despite Irregular Environmental Perturbations
Rhythmicity of coastal marine picoeukaryotes, bacteria and archaea despite irregular environmental perturbations Stefan Lambert, Margot Tragin, Jean-Claude Lozano, Jean-François Ghiglione, Daniel Vaulot, François-Yves Bouget, Pierre Galand To cite this version: Stefan Lambert, Margot Tragin, Jean-Claude Lozano, Jean-François Ghiglione, Daniel Vaulot, et al.. Rhythmicity of coastal marine picoeukaryotes, bacteria and archaea despite irregular environmental perturbations. ISME Journal, Nature Publishing Group, 2019, 13 (2), pp.388-401. 10.1038/s41396- 018-0281-z. hal-02326251 HAL Id: hal-02326251 https://hal.archives-ouvertes.fr/hal-02326251 Submitted on 19 Nov 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Rhythmicity of coastal marine picoeukaryotes, bacteria and archaea despite irregular environmental perturbations Stefan Lambert, Margot Tragin, Jean-Claude Lozano, Jean-François Ghiglione, Daniel Vaulot, François-Yves Bouget, Pierre Galand To cite this version: Stefan Lambert, Margot Tragin, Jean-Claude Lozano, Jean-François Ghiglione, Daniel -

Catalogue of Protozoan Parasites Recorded in Australia Peter J. O
1 CATALOGUE OF PROTOZOAN PARASITES RECORDED IN AUSTRALIA PETER J. O’DONOGHUE & ROBERT D. ADLARD O’Donoghue, P.J. & Adlard, R.D. 2000 02 29: Catalogue of protozoan parasites recorded in Australia. Memoirs of the Queensland Museum 45(1):1-164. Brisbane. ISSN 0079-8835. Published reports of protozoan species from Australian animals have been compiled into a host- parasite checklist, a parasite-host checklist and a cross-referenced bibliography. Protozoa listed include parasites, commensals and symbionts but free-living species have been excluded. Over 590 protozoan species are listed including amoebae, flagellates, ciliates and ‘sporozoa’ (the latter comprising apicomplexans, microsporans, myxozoans, haplosporidians and paramyxeans). Organisms are recorded in association with some 520 hosts including mammals, marsupials, birds, reptiles, amphibians, fish and invertebrates. Information has been abstracted from over 1,270 scientific publications predating 1999 and all records include taxonomic authorities, synonyms, common names, sites of infection within hosts and geographic locations. Protozoa, parasite checklist, host checklist, bibliography, Australia. Peter J. O’Donoghue, Department of Microbiology and Parasitology, The University of Queensland, St Lucia 4072, Australia; Robert D. Adlard, Protozoa Section, Queensland Museum, PO Box 3300, South Brisbane 4101, Australia; 31 January 2000. CONTENTS the literature for reports relevant to contemporary studies. Such problems could be avoided if all previous HOST-PARASITE CHECKLIST 5 records were consolidated into a single database. Most Mammals 5 researchers currently avail themselves of various Reptiles 21 electronic database and abstracting services but none Amphibians 26 include literature published earlier than 1985 and not all Birds 34 journal titles are covered in their databases. Fish 44 Invertebrates 54 Several catalogues of parasites in Australian PARASITE-HOST CHECKLIST 63 hosts have previously been published.