Pan-Prokaryote Ribopool Universal, Affordable Ribosomal RNA Depletion for Microbes (Bacteria/Archaea)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Diversity of Understudied Archaeal and Bacterial Populations of Yellowstone National Park: from Genes to Genomes Daniel Colman
University of New Mexico UNM Digital Repository Biology ETDs Electronic Theses and Dissertations 7-1-2015 Diversity of understudied archaeal and bacterial populations of Yellowstone National Park: from genes to genomes Daniel Colman Follow this and additional works at: https://digitalrepository.unm.edu/biol_etds Recommended Citation Colman, Daniel. "Diversity of understudied archaeal and bacterial populations of Yellowstone National Park: from genes to genomes." (2015). https://digitalrepository.unm.edu/biol_etds/18 This Dissertation is brought to you for free and open access by the Electronic Theses and Dissertations at UNM Digital Repository. It has been accepted for inclusion in Biology ETDs by an authorized administrator of UNM Digital Repository. For more information, please contact [email protected]. Daniel Robert Colman Candidate Biology Department This dissertation is approved, and it is acceptable in quality and form for publication: Approved by the Dissertation Committee: Cristina Takacs-Vesbach , Chairperson Robert Sinsabaugh Laura Crossey Diana Northup i Diversity of understudied archaeal and bacterial populations from Yellowstone National Park: from genes to genomes by Daniel Robert Colman B.S. Biology, University of New Mexico, 2009 DISSERTATION Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy Biology The University of New Mexico Albuquerque, New Mexico July 2015 ii DEDICATION I would like to dedicate this dissertation to my late grandfather, Kenneth Leo Colman, associate professor of Animal Science in the Wool laboratory at Montana State University, who even very near the end of his earthly tenure, thought it pertinent to quiz my knowledge of oxidized nitrogen compounds. He was a man of great curiosity about the natural world, and to whom I owe an acknowledgement for his legacy of intellectual (and actual) wanderlust. -

Ninety-Nine De Novo Assembled Genomes from the Moose (Alces Alces) Rumen Microbiome Provide New Insights Into Microbial Plant Biomass Degradation
The ISME Journal (2017) 11, 2538–2551 © 2017 International Society for Microbial Ecology All rights reserved 1751-7362/17 www.nature.com/ismej ORIGINAL ARTICLE Ninety-nine de novo assembled genomes from the moose (Alces alces) rumen microbiome provide new insights into microbial plant biomass degradation Olov Svartström1, Johannes Alneberg2, Nicolas Terrapon3,4, Vincent Lombard3,4, Ino de Bruijn2, Jonas Malmsten5,6, Ann-Marie Dalin6, Emilie EL Muller7, Pranjul Shah7, Paul Wilmes7, Bernard Henrissat3,4,8, Henrik Aspeborg1 and Anders F Andersson2 1School of Biotechnology, Division of Industrial Biotechnology, KTH Royal Institute of Technology, Stockholm, Sweden; 2School of Biotechnology, Division of Gene Technology, KTH Royal Institute of Technology, Science for Life Laboratory, Stockholm, Sweden; 3CNRS UMR 7257, Aix-Marseille University, 13288 Marseille, France; 4INRA, USC 1408 AFMB, 13288 Marseille, France; 5Department of Pathology and Wildlife Diseases, National Veterinary Institute, Uppsala, Sweden; 6Division of Reproduction, Department of Clinical Sciences, Swedish University of Agricultural Sciences, Uppsala, Sweden; 7Luxembourg Centre for Systems Biomedicine, University of Luxembourg, Esch-sur-Alzette, Luxembourg and 8Department of Biological Sciences, King Abdulaziz University, Jeddah, Saudi Arabia The moose (Alces alces) is a ruminant that harvests energy from fiber-rich lignocellulose material through carbohydrate-active enzymes (CAZymes) produced by its rumen microbes. We applied shotgun metagenomics to rumen contents from six moose to obtain insights into this microbiome. Following binning, 99 metagenome-assembled genomes (MAGs) belonging to 11 prokaryotic phyla were reconstructed and characterized based on phylogeny and CAZyme profile. The taxonomy of these MAGs reflected the overall composition of the metagenome, with dominance of the phyla Bacteroidetes and Firmicutes. -

Distribution and Diversity of Members of the Bacterial Phylum Fibrobacteres in Environments Where Cellulose Degradation Occurs
Aberystwyth University Distribution and diversity of members of the bacterial phylum Fibrobacteres in environments where cellulose degradation occurs. Ransom-Jones, Emma; Jones, Davey L.; Edwards, Arwyn; McDonald, J. E. Published in: Systematic and Applied Microbiology DOI: 10.1016/j.syapm.2014.06.001 Publication date: 2014 Citation for published version (APA): Ransom-Jones, E., Jones, D. L., Edwards, A., & McDonald, J. E. (2014). Distribution and diversity of members of the bacterial phylum Fibrobacteres in environments where cellulose degradation occurs. Systematic and Applied Microbiology, 37(7), 502-509. https://doi.org/10.1016/j.syapm.2014.06.001 General rights Copyright and moral rights for the publications made accessible in the Aberystwyth Research Portal (the Institutional Repository) are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. • Users may download and print one copy of any publication from the Aberystwyth Research Portal for the purpose of private study or research. • You may not further distribute the material or use it for any profit-making activity or commercial gain • You may freely distribute the URL identifying the publication in the Aberystwyth Research Portal Take down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. tel: +44 1970 62 2400 email: [email protected] Download date: 28. Sep. 2021 G Model SYAPM-25635; No. of Pages 8 ARTICLE IN PRESS Systematic and Applied Microbiology xxx (2014) xxx–xxx Contents lists available at ScienceDirect Systematic and Applied Microbiology j ournal homepage: www.elsevier.de/syapm Distribution and diversity of members of the bacterial phylum Fibrobacteres in environments where cellulose degradation occurs a b c a, Emma Ransom-Jones , David L. -

Expanding the Chlamydiae Tree
Digital Comprehensive Summaries of Uppsala Dissertations from the Faculty of Science and Technology 2040 Expanding the Chlamydiae tree Insights into genome diversity and evolution JENNAH E. DHARAMSHI ACTA UNIVERSITATIS UPSALIENSIS ISSN 1651-6214 ISBN 978-91-513-1203-3 UPPSALA urn:nbn:se:uu:diva-439996 2021 Dissertation presented at Uppsala University to be publicly examined in A1:111a, Biomedical Centre (BMC), Husargatan 3, Uppsala, Tuesday, 8 June 2021 at 13:15 for the degree of Doctor of Philosophy. The examination will be conducted in English. Faculty examiner: Prof. Dr. Alexander Probst (Faculty of Chemistry, University of Duisburg-Essen). Abstract Dharamshi, J. E. 2021. Expanding the Chlamydiae tree. Insights into genome diversity and evolution. Digital Comprehensive Summaries of Uppsala Dissertations from the Faculty of Science and Technology 2040. 87 pp. Uppsala: Acta Universitatis Upsaliensis. ISBN 978-91-513-1203-3. Chlamydiae is a phylum of obligate intracellular bacteria. They have a conserved lifecycle and infect eukaryotic hosts, ranging from animals to amoeba. Chlamydiae includes pathogens, and is well-studied from a medical perspective. However, the vast majority of chlamydiae diversity exists in environmental samples as part of the uncultivated microbial majority. Exploration of microbial diversity in anoxic deep marine sediments revealed diverse chlamydiae with high relative abundances. Using genome-resolved metagenomics various marine sediment chlamydiae genomes were obtained, which significantly expanded genomic sampling of Chlamydiae diversity. These genomes formed several new clades in phylogenomic analyses, and included Chlamydiaceae relatives. Despite endosymbiosis-associated genomic features, hosts were not identified, suggesting chlamydiae with alternate lifestyles. Genomic investigation of Anoxychlamydiales, newly described here, uncovered genes for hydrogen metabolism and anaerobiosis, suggesting they engage in syntrophic interactions. -

Phylogenetics of Archaeal Lipids Amy Kelly 9/27/2006 Outline
Phylogenetics of Archaeal Lipids Amy Kelly 9/27/2006 Outline • Phlogenetics of Archaea • Phlogenetics of archaeal lipids • Papers Phyla • Two? main phyla – Euryarchaeota • Methanogens • Extreme halophiles • Extreme thermophiles • Sulfate-reducing – Crenarchaeota • Extreme thermophiles – Korarchaeota? • Hyperthermophiles • indicated only by environmental DNA sequences – Nanoarchaeum? • N. equitans a fast evolving euryarchaeal lineage, not novel, early diverging archaeal phylum – Ancient archael group? • In deepest brances of Crenarchaea? Euryarchaea? Archaeal Lipids • Methanogens – Di- and tetra-ethers of glycerol and isoprenoid alcohols – Core mostly archaeol or caldarchaeol – Core sometimes sn-2- or Images removed due to sn-3-hydroxyarchaeol or copyright considerations. macrocyclic archaeol –PMI • Halophiles – Similar to methanogens – Exclusively synthesize bacterioruberin • Marine Crenarchaea Depositional Archaeal Lipids Biological Origin Environment Crocetane methanotrophs? methane seeps? methanogens, PMI (2,6,10,15,19-pentamethylicosane) methanotrophs hypersaline, anoxic Squalane hypersaline? C31-C40 head-to-head isoprenoids Smit & Mushegian • “Lost” enzymes of MVA pathway must exist – Phosphomevalonate kinase (PMK) – Diphosphomevalonate decarboxylase – Isopentenyl diphosphate isomerase (IPPI) Kaneda et al. 2001 Rohdich et al. 2001 Boucher et al. • Isoprenoid biosynthesis of archaea evolved through a combination of processes – Co-option of ancestral enzymes – Modification of enzymatic specificity – Orthologous and non-orthologous gene -

Global Metagenomic Survey Reveals a New Bacterial Candidate Phylum in Geothermal Springs
ARTICLE Received 13 Aug 2015 | Accepted 7 Dec 2015 | Published 27 Jan 2016 DOI: 10.1038/ncomms10476 OPEN Global metagenomic survey reveals a new bacterial candidate phylum in geothermal springs Emiley A. Eloe-Fadrosh1, David Paez-Espino1, Jessica Jarett1, Peter F. Dunfield2, Brian P. Hedlund3, Anne E. Dekas4, Stephen E. Grasby5, Allyson L. Brady6, Hailiang Dong7, Brandon R. Briggs8, Wen-Jun Li9, Danielle Goudeau1, Rex Malmstrom1, Amrita Pati1, Jennifer Pett-Ridge4, Edward M. Rubin1,10, Tanja Woyke1, Nikos C. Kyrpides1 & Natalia N. Ivanova1 Analysis of the increasing wealth of metagenomic data collected from diverse environments can lead to the discovery of novel branches on the tree of life. Here we analyse 5.2 Tb of metagenomic data collected globally to discover a novel bacterial phylum (‘Candidatus Kryptonia’) found exclusively in high-temperature pH-neutral geothermal springs. This lineage had remained hidden as a taxonomic ‘blind spot’ because of mismatches in the primers commonly used for ribosomal gene surveys. Genome reconstruction from metagenomic data combined with single-cell genomics results in several high-quality genomes representing four genera from the new phylum. Metabolic reconstruction indicates a heterotrophic lifestyle with conspicuous nutritional deficiencies, suggesting the need for metabolic complementarity with other microbes. Co-occurrence patterns identifies a number of putative partners, including an uncultured Armatimonadetes lineage. The discovery of Kryptonia within previously studied geothermal springs underscores the importance of globally sampled metagenomic data in detection of microbial novelty, and highlights the extraordinary diversity of microbial life still awaiting discovery. 1 Department of Energy Joint Genome Institute, Walnut Creek, California 94598, USA. 2 Department of Biological Sciences, University of Calgary, Calgary, Alberta T2N 1N4, Canada. -

Two Domains of Life, Not Three?
Asgard Archaea Two domains of life, not three? A picture of visual interpretation of the Wagner’s opera Das Rheingold, a part of Richard Wagner’s Der Ring des Nibelungen Loki's Castle is a field of five active hydrothermal vents in the mid-Atlantic Ocean, located at 73 degrees north on the Mid-Atlantic Ridge between Greenland and Norway at a depth of 2,352 metres The vents were discovered in 2008 by a multinational scientific expedition of the university of Bergen, and are the most northerly black smokers to date. The five active chimneys of Loki's Castle are venting water as hot as 300 °C and sit on a vast mound of sulfide minerals. The vent field was given the name Loki's Castle as its shape reminded its discoverers of a fantasy castle. The reference is to the ancient Norse god of trickery, Loki. The top three feet (1 m) of a vent chimney almost 40 feet (12 m) tall at Loki's Castle in mid-July 2008. Visible at left is the arm of a remotely operated vehicle, reaching in to take fluid samples. Loki’s Castle Wents Loki's Castle - a field of five active hydrothermal vents in the mid-Atlantic Ocean - at 73 degrees north on the Mid-Atlantic Ridge - at a depth of 2,352 meters In Norse mythology, Loki is a cunning trickster who has the ability to change his shape and sex. Loki is represented as the companion of the great gods Odin and Thor. Preliminary observations have shown the warm area around the Loki's Castle vents to be alive with diverse and apparently unique microorganisms, unlike vent communities observed elsewhere. -

Fiber-Associated Spirochetes Are Major Agents of Hemicellulose Degradation in the Hindgut of Wood-Feeding Higher Termites
Fiber-associated spirochetes are major agents of hemicellulose degradation in the hindgut of wood-feeding higher termites Gaku Tokudaa,b,1, Aram Mikaelyanc,d, Chiho Fukuia, Yu Matsuuraa, Hirofumi Watanabee, Masahiro Fujishimaf, and Andreas Brunec aTropical Biosphere Research Center, Center of Molecular Biosciences, University of the Ryukyus, Nishihara, 903-0213 Okinawa, Japan; bGraduate School of Engineering and Science, University of the Ryukyus, Nishihara, 903-0213 Okinawa, Japan; cResearch Group Insect Gut Microbiology and Symbiosis, Max Planck Institute for Terrestrial Microbiology, 35043 Marburg, Germany; dDepartment of Entomology and Plant Pathology, North Carolina State University, Raleigh, NC 27607; eBiomolecular Mimetics Research Unit, Institute of Agrobiological Sciences, National Agriculture and Food Research Organization, Tsukuba, 305-8634 Ibaraki, Japan; and fDepartment of Sciences, Graduate School of Sciences and Technology for Innovation, Yamaguchi University, Yoshida 1677-1, 753-8512 Yamaguchi, Japan Edited by Nancy A. Moran, University of Texas at Austin, Austin, TX, and approved November 5, 2018 (received for review June 25, 2018) Symbiotic digestion of lignocellulose in wood-feeding higher digestion in the hindgut of higher termites must be attributed to termites (family Termitidae) is a two-step process that involves their entirely prokaryotic microbial community (5). endogenous host cellulases secreted in the midgut and a dense The gut microbiota of higher termites comprises more than bacterial community in the hindgut compartment. The genomes of 1,000 bacterial phylotypes, which are organized into distinc- the bacterial gut microbiota encode diverse cellulolytic and hemi- tive communities colonizing the microhabitats provided by the cellulolytic enzymes, but the contributions of host and bacterial compartmentalized intestine, including the highly differentiated symbionts to lignocellulose degradation remain ambiguous. -

Archaeology of Eukaryotic DNA Replication
Downloaded from http://cshperspectives.cshlp.org/ on September 25, 2021 - Published by Cold Spring Harbor Laboratory Press Archaeology of Eukaryotic DNA Replication Kira S. Makarova and Eugene V. Koonin National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, Maryland 20894 Correspondence: [email protected] Recent advances in the characterization of the archaeal DNA replication system together with comparative genomic analysis have led to the identification of several previously un- characterized archaeal proteins involved in replication and currently reveal a nearly com- plete correspondence between the components of the archaeal and eukaryotic replication machineries. It can be inferred that the archaeal ancestor of eukaryotes and even the last common ancestor of all extant archaea possessed replication machineries that were compa- rable in complexity to the eukaryotic replication system. The eukaryotic replication system encompasses multiple paralogs of ancestral components such that heteromeric complexes in eukaryotes replace archaeal homomeric complexes, apparently along with subfunctionali- zation of the eukaryotic complex subunits. In the archaea, parallel, lineage-specific dupli- cations of many genes encoding replication machinery components are detectable as well; most of these archaeal paralogs remain to be functionally characterized. The archaeal rep- lication system shows remarkable plasticity whereby even some essential components such as DNA polymerase and single-stranded DNA-binding protein are displaced by unrelated proteins with analogous activities in some lineages. ouble-stranded DNA is the molecule that Okazaki fragments (Kornberg and Baker 2005; Dcarries genetic information in all cellular Barry and Bell 2006; Hamdan and Richardson life-forms; thus, replication of this genetic ma- 2009; Hamdan and van Oijen 2010). -
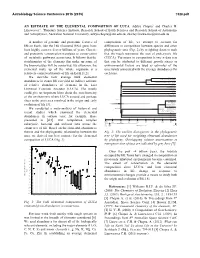
An Estimate of the Elemental Composition of Luca
Astrobiology Science Conference 2015 (2015) 7328.pdf AN ESTIMATE OF THE ELEMENTAL COMPOSITION OF LUCA. Aditya Chopra1 and Charles H. Lineweaver1, 1Planetary Science Institute, Research School of Earth Sciences and Research School of Astronomy and Astrophysics, Australian National University, [email protected], [email protected] A number of genomic and proteomic features of composition of life, we attempt to account for life on Earth, like the 16S ribosomal RNA gene, have differences in composition between species and other been highly conserved over billions of years. Genetic phylogenetic taxa (Fig. 2) by weighting datasets such and proteomic conservation translates to conservation that the result represents the root of prokaryotic life of metabolic pathways across taxa. It follows that the (LUCA). Variations in composition between data sets stoichiometry of the elements that make up some of that can be attributed to different growth stages or the biomolecules will be conserved. By extension, the environmental factors are used as estimates of the elemental make up of the whole organism is a uncertainty associated with the average abundances for relatively conserved feature of life on Earth [1,2]. each taxa. We describe how average bulk elemental Euryarchaeota 1a Methanococci, Methanobacteria, Methanopyri 7 Euryarchaeota 1b abundances in extant life can yield an indirect estimate 6 Thermoplasmata, Methanomicrobia, Halobacteria, Archaeoglobi Euryarchaeota 2 4 Thermococci of relative abundances of elements in the Last Crenarchaeota Sulfolobus, Thermoproteus ? Thaumarchaeota Universal Common Ancestor (LUCA). The results Cenarchaeum ? Korarchaeota ARMAN could give us important hints about the stoichiometry ? 2 Archaeal Richmond Mine Acidophilic Nanoorganisms Nanoarchaeota of the environment where LUCA existed and perhaps Archaea Terrabacteria clues to the processes involved in the origin and early Actinobacteria, Deinococcus-Thermus, 1 Cyanobacteria, Life Chloroflexi, 9 evolution of life [3]. -

Functionalized Membrane Domains: an Ancestral Feature of Archaea? Maxime Tourte, Philippe Schaeffer, Vincent Grossi, Phil Oger
Functionalized Membrane Domains: An Ancestral Feature of Archaea? Maxime Tourte, Philippe Schaeffer, Vincent Grossi, Phil Oger To cite this version: Maxime Tourte, Philippe Schaeffer, Vincent Grossi, Phil Oger. Functionalized Membrane Domains: An Ancestral Feature of Archaea?. Frontiers in Microbiology, Frontiers Media, 2020, 11, pp.526. 10.3389/fmicb.2020.00526. hal-02553764 HAL Id: hal-02553764 https://hal.archives-ouvertes.fr/hal-02553764 Submitted on 20 May 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. fmicb-11-00526 March 30, 2020 Time: 21:44 # 1 ORIGINAL RESEARCH published: 31 March 2020 doi: 10.3389/fmicb.2020.00526 Functionalized Membrane Domains: An Ancestral Feature of Archaea? Maxime Tourte1†, Philippe Schaeffer2†, Vincent Grossi3† and Phil M. Oger1*† 1 Université de Lyon, INSA Lyon, CNRS, MAP UMR 5240, Villeurbanne, France, 2 Université de Strasbourg-CNRS, UMR 7177, Laboratoire de Biogéochimie Moléculaire, Strasbourg, France, 3 Université de Lyon, ENS Lyon, CNRS, Laboratoire de Géologie de Lyon, UMR 5276, Villeurbanne, France Bacteria and Eukarya organize their plasma membrane spatially into domains of distinct functions. Due to the uniqueness of their lipids, membrane functionalization in Archaea remains a debated area. -

Universidad Politécnica De Madrid Escuela Técnica Superior De Ingeniería Agronómica Alimentaria Y De Biosistemas Departamento De Biotecnología-Biología Vegetal
UNIVERSIDAD POLITÉCNICA DE MADRID ESCUELA TÉCNICA SUPERIOR DE INGENIERÍA AGRONÓMICA ALIMENTARIA Y DE BIOSISTEMAS DEPARTAMENTO DE BIOTECNOLOGÍA-BIOLOGÍA VEGETAL TESIS DOCTORAL Caracterización de una nueva proteína hipertermófila NifB y su papel en el mecanismo de síntesis de NifB-co, precursor del grupo metálico de FeMo-co Author: Alessandro Scandurra Director: Prof. Luis Manuel Rubio Herrero Co-director: Dr. Simon Jean Marius Arragain i ii UNIVERSIDAD POLITÉCNICA DE MADRID ESCUELA TÉCNICA SUPERIOR DE INGENIERÍA AGRONÓMICA ALIMENTARIA Y DE BIOSISTEMAS DEPARTAMENTO DE BIOTECNOLOGÍA-BIOLOGÍA VEGETAL Memoria presentada por D. Alessandro Scandurra para optar al grado de Doctor Director Dr. Luis Manuel Rubio Herrero Profesor Titular UPM Madrid, 2017 © This copy oF the thesis has been supplied on condition that anyone who consults it is understood to recognize that its copyright rest with the author and that no quotation From the thesis, or any inFormation derived therefrom may be published without the author’s prior, written consent. I II AcKnowledgments As each journey, also this one has an end (Thanks goodness). It was a long trip, with several changes oF direction. Like For many other people, the PhD adventure was not easy, but I have to admit that I was lucky enough to meet really unique people, that became special Friendships. First oF all, I want to thank my Family, especially my mother and my Father, who always push me to go Forward in my life and to Follow my dreams, ready to pay the sacrifice oF the distance. This expirience has helped me to understand how easy is to be a son and how challenging is to be a parent.