Title Phylogenetic Distribution and Expression Pattern Analyses
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Complete Plastome Sequences Of
Karol et al. BMC Evolutionary Biology 2010, 10:321 http://www.biomedcentral.com/1471-2148/10/321 RESEARCH ARTICLE Open Access Complete plastome sequences of Equisetum arvense and Isoetes flaccida: implications for phylogeny and plastid genome evolution of early land plant lineages Kenneth G Karol1*, Kathiravetpillai Arumuganathan2, Jeffrey L Boore3,4, Aaron M Duffy5, Karin DE Everett6, John D Hall1, S Kellon Hansen5, Jennifer V Kuehl7, Dina F Mandoli6,8, Brent D Mishler9, Richard G Olmstead6, Karen S Renzaglia10, Paul G Wolf5 Abstract Background: Despite considerable progress in our understanding of land plant phylogeny, several nodes in the green tree of life remain poorly resolved. Furthermore, the bulk of currently available data come from only a subset of major land plant clades. Here we examine early land plant evolution using complete plastome sequences including two previously unexamined and phylogenetically critical lineages. To better understand the evolution of land plants and their plastomes, we examined aligned nucleotide sequences, indels, gene and nucleotide composition, inversions, and gene order at the boundaries of the inverted repeats. Results: We present the plastome sequences of Equisetum arvense, a horsetail, and of Isoetes flaccida,a heterosporous lycophyte. Phylogenetic analysis of aligned nucleotides from 49 plastome genes from 43 taxa supported monophyly for the following clades: embryophytes (land plants), lycophytes, monilophytes (leptosporangiate ferns + Angiopteris evecta + Psilotum nudum + Equisetum arvense), and seed plants. Resolution among the four monilophyte lineages remained moderate, although nucleotide analyses suggested that P. nudum and E. arvense form a clade sister to A. evecta + leptosporangiate ferns. Results from phylogenetic analyses of nucleotides were consistent with the distribution of plastome gene rearrangements and with analysis of sequence gaps resulting from insertions and deletions (indels). -

Botany Without Bias
editorial Botany without bias In the Gospel According to Matthew Chapter seven, Verse fve, Jesus says “frst cast out the beam out of thine own eye; and then shalt thou see clearly to cast out the mote out of thy brother’s eye”. We should remember this entreaty before too casually casting accusations of ‘plant blindness’. anguage usage helps maintain unconscious biases. In plant biology Lfor example, there is the careless use of the term ‘higher plants’, without thinking about its meaning or implication. If there is a definition of ‘higher plants’ then it is synonymous with vascular plants, but the image it conjures is of upstanding, leafy land-dwelling plants. The problem is that ‘higher’ is a charged term implying superiority of this group over their non-vascular cousins. This stratification is a manifestation of orthogenesis, the idea that evolution has both a direction and a goal. A perfect illustration of orthogenesis is the frequent meme of The Road to Homo Sapiens, the original version of which was drawn by Rudolph Zallinger for a 1965 edition of Life Nature Library1. Also known as The March of Progress, it shows a line of assumed human ancestors, starting with a gibbon-like in their News and Views3, “innovations spend much of their discussions on what Pliopithecus, processing from left to right, associated with improving water use the similarities of these organisms to becoming taller and more upright in stance, efficiency […] may be more fundamental angiosperms can tell about the history and culminating in a modern human. to the evolution of vascular plants than the of plants’ colonization of dry land, and The implication is clear, our evolutionary vascular system from which they derive much less on their characteristics and ancestors are only of interest as waypoints their name”. -
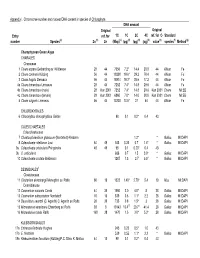
Green Appendix I
Appendix I. Chromsome number and nuclear DNA content in species of Chlorophyta DNA amount Original Original Entry ref. for 1C 1C 2C 4C ref. for C- Standard number Species(a) 2n (b) 2n (Mbp)(c) (pg)(d) (pg)(d) (pg)(d) value(e) species(f) Method(g) Charophycean Green Algae CHARALES Characeae 1 Chara aspera Detharding ex Willdenow 28 44 7056 7.2* 14.4 28.8 44 Allium Fe 2 Chara contraria Kützing 56 44 19208 19.6* 39.2 78.4 44 Allium Fe 3 Chara fragilis Desvaux 56 44 18914 19.3* 38.6 77.2 44 Allium Fe 4a Chara tomentosa Linnaeus 28 44 7252 7.4* 14.8 29.6 44 Allium Fe 4b Chara tomentosa (male) 28 Kun 2001 7252 7.4* 14.8 29.6 Kun 2001 Chara MI:EB 4c Chara tomentsoa (female) 28 Kun 2001 6860 7.0* 14.0 28.0 Kun 2001 Chara MI:EB 5 Chara vulgaris Linnaeus 56 44 13230 13.5* 27 54 44 Allium Fe CHLOROKYBALES 6 Chlorokybus atmosphyticus Geitler 98 0.1 0.2* 0.4 43 COLEOCHAETALES Coleochaetaceae 7 Chaetosphaeridium globosum (Nordstedt) Klebahn 1.2* * Gallus MI:DAPI 8 Coleochaete nitellarum Jost 84 49 343 0.35 0.7 1.4* * Gallus MI:DAPI 9a Coleochaete orbicularis Pringsheim 48 49 98 0.1 0.20* 0.4 43 9b C. orbicularis 686 0.7 1.5 3.0* * Gallus MI:DAPI 10 Coleochaete scutata Brébisson 1287 1.3 2.7 5.5* * Gallus MI:DAPI DESMIDIALES1 Closteriaceae 11 Closterium ehrenbergii Meneghini ex Ralfs 60 19 1323 1.40* 2.70* 5.4 19 Mus MI:DAPI Desmidiaceae 12 Cosmarium cucumis Corda 44 39 1960 2.0 4.0* 8 28 Gallus MI:DAPI 13 Cosmarium subcostatum Nordstedt 10 16 539 0.6 1.1* 2.2 28 Gallus MI:DAPI 14 Desmidium swartzii (C. -

From Algae to Flowering Plants
PP62CH23-Popper ARI 4 April 2011 14:20 Evolution and Diversity of Plant Cell Walls: From Algae to Flowering Plants Zoe¨ A. Popper,1 Gurvan Michel,3,4 Cecile´ Herve,´ 3,4 David S. Domozych,5 William G.T. Willats,6 Maria G. Tuohy,2 Bernard Kloareg,3,4 and Dagmar B. Stengel1 1Botany and Plant Science, and 2Molecular Glycotechnology Group, Biochemistry, School of Natural Sciences, National University of Ireland, Galway, Ireland; email: [email protected] 3CNRS and 4UPMC University Paris 6, UMR 7139 Marine Plants and Biomolecules, Station Biologique de Roscoff, F-29682 Roscoff, Bretagne, France 5Department of Biology and Skidmore Microscopy Imaging Center, Skidmore College, Saratoga Springs, New York 12866 6Department of Plant Biology and Biochemistry, Faculty of Life Sciences, University of Copenhagen, Bulowsvej,¨ 17-1870 Frederiksberg, Denmark Annu. Rev. Plant Biol. 2011. 62:567–90 Keywords First published online as a Review in Advance on xyloglucan, mannan, arabinogalactan proteins, genome, environment, February 22, 2011 multicellularity The Annual Review of Plant Biology is online at plant.annualreviews.org Abstract This article’s doi: All photosynthetic multicellular Eukaryotes, including land plants 10.1146/annurev-arplant-042110-103809 by Universidad Veracruzana on 01/08/14. For personal use only. and algae, have cells that are surrounded by a dynamic, complex, Copyright c 2011 by Annual Reviews. carbohydrate-rich cell wall. The cell wall exerts considerable biologi- All rights reserved cal and biomechanical control over individual cells and organisms, thus Annu. Rev. Plant Biol. 2011.62:567-590. Downloaded from www.annualreviews.org 1543-5008/11/0602-0567$20.00 playing a key role in their environmental interactions. -

The Chloroplast and Mitochondrial Genome Sequences of The
The chloroplast and mitochondrial genome sequences of the charophyte Chaetosphaeridium globosum: Insights into the timing of the events that restructured organelle DNAs within the green algal lineage that led to land plants Monique Turmel*, Christian Otis, and Claude Lemieux Canadian Institute for Advanced Research, Program in Evolutionary Biology, and De´partement de Biochimie et de Microbiologie, Universite´Laval, Que´bec, QC, Canada G1K 7P4 Edited by Jeffrey D. Palmer, Indiana University, Bloomington, IN, and approved June 19, 2002 (received for review April 5, 2002) The land plants and their immediate green algal ancestors, the organelle DNAs (mainly for the cpDNA of Spirogyra maxima,a charophytes, form the Streptophyta. There is evidence that both member of the Zygnematales; refs. 11–13), it is difficult to predict the chloroplast DNA (cpDNA) and mitochondrial DNA (mtDNA) the timing of the major events that shaped the architectures of underwent substantial changes in their architecture (intron inser- land-plant cpDNAs and mtDNAs. tions, gene losses, scrambling in gene order, and genome expan- Mesostigma cpDNA is highly similar in size and gene organization sion in the case of mtDNA) during the evolution of streptophytes; to the cpDNAs of the 10 photosynthetic land plants examined thus however, because no charophyte organelle DNAs have been se- far (the bryophyte Marchantia polymorpha and nine vascular plants; quenced completely thus far, the suite of events that shaped see www.ncbi.nlm.nih.gov͞PMGifs͞Genomes͞organelles.html), streptophyte organelle genomes remains largely unknown. Here, but lacks any introns and contains Ϸ20 extra genes (7). Chloroplast we have determined the complete cpDNA (131,183 bp) and mtDNA gene loss is an ongoing process in the Streptophyta, with indepen- (56,574 bp) sequences of the charophyte Chaetosphaeridium glo- dent losses occurring in multiple lineages (14, 15). -

Phylotranscriptomic Analysis of the Origin and Early PNAS PLUS Diversification of Land Plants
Phylotranscriptomic analysis of the origin and early PNAS PLUS diversification of land plants Norman J. Wicketta,b,1,2, Siavash Mirarabc,1, Nam Nguyenc, Tandy Warnowc, Eric Carpenterd, Naim Matascie,f, Saravanaraj Ayyampalayamg, Michael S. Barkerf, J. Gordon Burleighh, Matthew A. Gitzendannerh,i, Brad R. Ruhfelh,j,k, Eric Wafulal, Joshua P. Derl, Sean W. Grahamm, Sarah Mathewsn, Michael Melkoniano, Douglas E. Soltish,i,k, Pamela S. Soltish,i,k, Nicholas W. Milesk, Carl J. Rothfelsp,q, Lisa Pokornyp,r, A. Jonathan Shawp, Lisa DeGironimos, Dennis W. Stevensons, Barbara Sureko, Juan Carlos Villarrealt,BéatriceRoureu, Hervé Philippeu,v, Claude W. dePamphilisl, Tao Chenw, Michael K. Deyholosd, Regina S. Baucomx, Toni M. Kutchany, Megan M. Augustiny,JunWangz, Yong Zhangv, Zhijian Tianz,ZhixiangYanz,XiaoleiWuz,XiaoSunz, Gane Ka-Shu Wongd,z,aa,2, and James Leebens-Mackg,2 aChicago Botanic Garden, Glencoe, IL 60022; bProgram in Biological Sciences, Northwestern University, Evanston, IL 60208; cDepartment of Computer Science, University of Texas, Austin, TX 78712; dDepartment of Biological Sciences, University of Alberta, Edmonton, AB, Canada T6G 2E9; eiPlant Collaborative, Tucson, AZ 85721; fDepartment of Ecology and Evolutionary Biology, University of Arizona, Tucson, AZ 85721; gDepartment of Plant Biology, University of Georgia, Athens, GA 30602; hDepartment of Biology and iGenetics Institute, University of Florida, Gainesville, FL 32611; jDepartment of Biological Sciences, Eastern Kentucky University, Richmond, KY 40475; kFlorida Museum of -

Embryophyta - Jean Broutin
PHYLOGENETIC TREE OF LIFE – Embryophyta - Jean Broutin EMBRYOPHYTA Jean Broutin UMR 7207, CNRS/MNHN/UPMC, Sorbonne Universités, UPMC, Paris Keywords: Embryophyta, phylogeny, classification, morphology, green plants, land plants, lineages, diversification, life history, Bryophyta, Tracheophyta, Spermatophyta, gymnosperms, angiosperms. Contents 1. Introduction to land plants 2. Embryophytes. Characteristics and diversity 2.1. Human Use of Embryophytes 2.2. Importance of the Embryophytes in the History of Life 2.3. Phylogenetic Emergence of the Embryophytes: The “Streptophytes” Concept 2.4. Evolutionary Origin of the Embryophytes: The Phyletic Lineages within the Embryophytes 2.5. Invasion of Land and Air by the Embryophytes: A Complex Evolutionary Success 2.6. Hypotheses about the First Appearance of Embryophytes, the Fossil Data 3. Bryophytes 3.1. Division Marchantiophyta (liverworts) 3.2. Division Anthocerotophyta (hornworts) 3.3. Division Bryophyta (mosses) 4. Tracheophytes (vascular plants) 4.1. The “Polysporangiophyte Concept” and the Tracheophytes 4.2. Tracheophyta (“true” Vascular Plants) 4.3. Eutracheophyta 4.3.1. Seedless Vascular Plants 4.3.1.1. Rhyniophyta (Extinct Group) 4.3.1.2. Lycophyta 4.3.1.3. Euphyllophyta 4.3.1.4. Monilophyta 4.3.1.5. Eusporangiate Ferns 4.3.1.6. Psilotales 4.3.1.7. Ophioglossales 4.3.1.8. Marattiales 4.3.1.9. Equisetales 4.3.1.10. Leptosporangiate Ferns 4.3.1.11. Polypodiales 4.3.1.12. Cyatheales 4.3.1.13. Salviniales 4.3.1.14. Osmundales 4.3.1.15. Schizaeales, Gleicheniales, Hymenophyllales. 5. Progymnosperm concept: the “emblematic” fossil plant Archaeopteris. 6. Seed plants 6.1. Spermatophyta 6.1.1. Gymnosperms ©Encyclopedia of Life Support Systems (EOLSS) PHYLOGENETIC TREE OF LIFE – Embryophyta - Jean Broutin 6.1.2. -

Phylotranscriptomic Analysis of the Origin and Early Diversification Of
Phylotranscriptomic analysis of the origin and early PNAS PLUS diversification of land plants Norman J. Wicketta,b,1,2, Siavash Mirarabc,1, Nam Nguyenc, Tandy Warnowc, Eric Carpenterd, Naim Matascie,f, Saravanaraj Ayyampalayamg, Michael S. Barkerf, J. Gordon Burleighh, Matthew A. Gitzendannerh,i, Brad R. Ruhfelh,j,k, Eric Wafulal, Joshua P. Derl, Sean W. Grahamm, Sarah Mathewsn, Michael Melkoniano, Douglas E. Soltish,i,k, Pamela S. Soltish,i,k, Nicholas W. Milesk, Carl J. Rothfelsp,q, Lisa Pokornyp,r, A. Jonathan Shawp, Lisa DeGironimos, Dennis W. Stevensons, Barbara Sureko, Juan Carlos Villarrealt,BéatriceRoureu, Hervé Philippeu,v, Claude W. dePamphilisl, Tao Chenw, Michael K. Deyholosd, Regina S. Baucomx, Toni M. Kutchany, Megan M. Augustiny,JunWangz, Yong Zhangv, Zhijian Tianz,ZhixiangYanz,XiaoleiWuz,XiaoSunz, Gane Ka-Shu Wongd,z,aa,2, and James Leebens-Mackg,2 aChicago Botanic Garden, Glencoe, IL 60022; bProgram in Biological Sciences, Northwestern University, Evanston, IL 60208; cDepartment of Computer Science, University of Texas, Austin, TX 78712; dDepartment of Biological Sciences, University of Alberta, Edmonton, AB, Canada T6G 2E9; eiPlant Collaborative, Tucson, AZ 85721; fDepartment of Ecology and Evolutionary Biology, University of Arizona, Tucson, AZ 85721; gDepartment of Plant Biology, University of Georgia, Athens, GA 30602; hDepartment of Biology and iGenetics Institute, University of Florida, Gainesville, FL 32611; jDepartment of Biological Sciences, Eastern Kentucky University, Richmond, KY 40475; kFlorida Museum of -

Evolutionary Transitions Between States of Structural and Developmental Characters Among the Algal Charophyta (Viridiplantae)
ABSTRACT Title of Thesis: EVOLUTIONARY TRANSITIONS BETWEEN STATES OF STRUCTURAL AND DEVELOPMENTAL CHARACTERS AMONG THE ALGAL CHAROPHYTA (VIRIDIPLANTAE). Jeffrey David Lewandowski, Master of Science, 2004 Thesis Directed By: Associate Profe ssor, Charles F. Delwiche, Cell Biology and Molecular Genetics The Charophyta comprise green plant representatives ranging from familiar complex -bodied land plants to subtle and simple forms of green algae, presumably the closest phylogenetic relatives of land plants. This biological lineage provides a unique opportunity to investigate evolutionary transition series that likely facilitated once -aquatic green plants to colonize and diversify in terrestrial environments. A literature review summarizes fu ndamental structural and developmental transitions observed among the major lineages of algal Charophyta. A phylogenetic framework independent of morphological and ontological data is necessary for testing hypotheses about the evolution of structure and development. Thus, to further elucidate the branching order of the algal Charophyta, new DNA sequence data are used to test conflicting hypotheses regarding the phylogenetic placement of several enigmatic taxa, including the algal charophyte genera Mesosti gma , Chlorokybus, Coleochaete , and Chaetosphaeridium. Additionally, technical notes on developing RNA methods for use in studying algal Charophyta are included. EVOLUTIONARY TRANSITIONS BETWEEN STATES OF STRUCTURAL AND DEVELOPMENTAL CHARACTERS AMONG THE ALGAL CHAROPHYTA (VIRIDIPLANTAE). By Jeffrey David Lewandowski Thesis submitted to the Faculty of the Graduate School of the University of Maryland, College Park, in partial fulfillment of the requirements for the degree of Master of Science 2004 Advisory Committee: Associate Professor Charles F. Delwiche, Chair Professor Todd J. Cooke Assistant Professor Eric S. Haag © Copyright by Jeffrey D. Lewandowski 2004 Preface The following are examples from several of the works that have provided philosophical inspiration for this document. -

Basket Scales of the Green Alga, Mesostigma Viride: Chemistry and Ultrastructure
Basket scales of the green alga, Mesostigma viride: chemistry and ultrastructure DAVID S. DOMOZYCH* Department of Biology, Skidmore College, Saratoga Springs, NY 12866, USA BRIAN WELLS and PETER J. SHAW Department of Cell Biology, John Innes Institute for Plant Science Research, Colney Lane, Norwich NR4 7UH, UK * Author for correspondence Summary The unicellular green algal flagellate, Mesostigma tially digested scales revealed the presence of two viride, possesses an extracellular matrix consisting protein components, a high Mr, non-migrating form of three layers of highly distinct scales. This study and a form with a Mr of approximately 48000. focused upon the elaborate basket scales. The basket Further biochemical analyses of scales showed the scale is approximately 450-500 nm in height and presence of two major sugars: glucose and the consists of a solid base, two distinct lattices, two unusual keto sugar acid, 3-deoxy-lyxo-2-heptulosaric upper rims and various struts and roots. Pure acid, or DHA. Antibodies to scales were raised in rats preparations of isolated basket scales were obtained and used in immunolabeling studies. It is estimated and used for subsequent chemical and immunologi- from immunofluorescence studies that a cell of 8 /on cal analyses. X-ray microanalyses revealed the scale contains about 800 basket scales. The scales are in a as being mineralized with both calcium and phos- close-packed arrangement resulting in quasi-crystal- phorus. Fourier Transform Infrared Spectroscopic line arrays upon the cell surface. analyses revealed the presence of phosphate, sugges- ting in turn that the basket scale is complexed with Key words: basket scales, extracellular matrix, Mesostigma, calcium phosphate. -
Angiosperm Phylogeny Inferred from Sequences of Four Mitochondrial Genes 1Yin-Long QIU∗ 1Libo LI 1Bin WANG 1,2Jia-Yu XUE 1Tory A
Journal of Systematics and Evolution 48 (6): 391–425 (2010) doi: 10.1111/j.1759-6831.2010.00097.x Angiosperm phylogeny inferred from sequences of four mitochondrial genes 1Yin-Long QIU∗ 1Libo LI 1Bin WANG 1,2Jia-Yu XUE 1Tory A. HENDRY 1Rui-Qi LI 1Joseph W. BROWN 1Ya n g L I U 1Geordan T. HUDSON 3Zhi-Duan CHEN 1(Department of Ecology and Evolutionary Biology, University of Michigan, Ann Arbor, MI 48109, USA) 2(School of Life Sciences, Nanjing University, Nanjing 210093, China) 3(Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China) Abstract An angiosperm phylogeny was reconstructed in a maximum likelihood analysis of sequences of four mitochondrial genes, atp1, matR, nad5, and rps3, from 380 species that represent 376 genera and 296 families of seed plants. It is largely congruent with the phylogeny of angiosperms reconstructed from chloroplast genes atpB, matK, and rbcL, and nuclear 18S rDNA. The basalmost lineage consists of Amborella and Nymphaeales (including Hydatellaceae). Austrobaileyales follow this clade and are sister to the mesangiosperms, which include Chloranthaceae, Ceratophyllum, magnoliids, monocots, and eudicots. With the exception of Chloranthaceae being sister to Ceratophyllum, relationships among these five lineages are not well supported. In eudicots, Ranunculales, Sabiales, Proteales, Trochodendrales, Buxales, Gunnerales, Saxifragales, Vitales, Berberidopsidales, and Dilleniales form a basal grade of lines that diverged before the diversification of rosids and asterids. Within rosids, the COM (Celastrales–Oxalidales–Malpighiales) clade is sister to malvids (or rosid II), instead of to the nitrogen-fixing clade as found in all previous large-scale molecular analyses of angiosperms. Santalales and Caryophyllales are members of an expanded asterid clade. -
The Origin of Plants: Body Plan Changes Contributing to a Major Evolutionary Radiation
Review The origin of plants: Body plan changes contributing to a major evolutionary radiation Linda E. Graham*†, Martha E. Cook‡, and James S. Busse§ *Department of Botany, University of Wisconsin, Madison, WI 53706-1381; ‡Department of Biological Sciences, Illinois State University, Normal, IL 61790-4120; §Department of Biochemistry, University of Wisconsin, Madison, WI 53706-1381 Edited by Andrew H. Knoll, Harvard University, Cambridge, MA, and approved February 22, 2000 (received for review January 10, 2000) lants (land plants, embryophytes) are of monophyletic origin considered to be early divergent and not in the green algal Pfrom a freshwater ancestor that, if still extant, would be ancestors of plants. Early divergent plant forms include bryo- classified among the charophycean green algae. Plants, but not phytes (liverworts, hornworts, mosses), simple rootless plants charophyceans, possess a life history involving alternation of two considered to lack specialized food and lignified water conduct- morphologically distinct developmentally associated bodies, ing cells (vascular tissue), as well as simple vascular plants that sporophyte and gametophyte. Body plan evolution in plants has produce spores rather than seeds. Later-appearing vascular plant involved fundamental changes in the forms of both gametophyte groups are characterized by multicellular shoot and root apical and sporophyte and the evolutionary origin of regulatory sys- meristems. Vascular plants differ from bryophytes in possessing tems that generate different body plans in sporophytes and two types of apical meristem, those of the shoot and root. Woody gametophytes of the same species. Comparative analysis, based vascular plants possess two additional forms of meristematic on molecular phylogenetic information, identifies fundamental tissues, the vascular and cork cambia.