The Chloroplast and Mitochondrial Genome Sequences of The
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Complete Plastome Sequences Of
Karol et al. BMC Evolutionary Biology 2010, 10:321 http://www.biomedcentral.com/1471-2148/10/321 RESEARCH ARTICLE Open Access Complete plastome sequences of Equisetum arvense and Isoetes flaccida: implications for phylogeny and plastid genome evolution of early land plant lineages Kenneth G Karol1*, Kathiravetpillai Arumuganathan2, Jeffrey L Boore3,4, Aaron M Duffy5, Karin DE Everett6, John D Hall1, S Kellon Hansen5, Jennifer V Kuehl7, Dina F Mandoli6,8, Brent D Mishler9, Richard G Olmstead6, Karen S Renzaglia10, Paul G Wolf5 Abstract Background: Despite considerable progress in our understanding of land plant phylogeny, several nodes in the green tree of life remain poorly resolved. Furthermore, the bulk of currently available data come from only a subset of major land plant clades. Here we examine early land plant evolution using complete plastome sequences including two previously unexamined and phylogenetically critical lineages. To better understand the evolution of land plants and their plastomes, we examined aligned nucleotide sequences, indels, gene and nucleotide composition, inversions, and gene order at the boundaries of the inverted repeats. Results: We present the plastome sequences of Equisetum arvense, a horsetail, and of Isoetes flaccida,a heterosporous lycophyte. Phylogenetic analysis of aligned nucleotides from 49 plastome genes from 43 taxa supported monophyly for the following clades: embryophytes (land plants), lycophytes, monilophytes (leptosporangiate ferns + Angiopteris evecta + Psilotum nudum + Equisetum arvense), and seed plants. Resolution among the four monilophyte lineages remained moderate, although nucleotide analyses suggested that P. nudum and E. arvense form a clade sister to A. evecta + leptosporangiate ferns. Results from phylogenetic analyses of nucleotides were consistent with the distribution of plastome gene rearrangements and with analysis of sequence gaps resulting from insertions and deletions (indels). -

Evolution and Comparative Morphology of the Euglenophyte
EVOLUTION AND COMPARATIVE MORPHOLOGY OF THE EUGLENOPHYTE PLASTID by PATRICK JERRY PAUL BROWN (Under the Direction of Mark A. Farmer) ABSTRACT My doctoral research centered on understanding the evolution of the euglenophyte protists, with special attention paid to their plastids. The euglenophytes are a widely distributed group of euglenid protists that have acquired a chloroplast via secondary symbiogenesis. The goals of my research were to 1) test the efficacy of plastid morphological and ultrastructural characters in phylogenetic analysis; 2) understand the process of plastid development and partitioning in the euglenophytes; 3) to use a plastid- encoded protein gene to determine a euglenophyte phylogeny; and 4) to perform a multi- gene analysis to uncover clues about the origins of the euglenophyte plastid. My work began with an alpha-taxonomic study that redefined the rare euglenophyte Euglena rustica. This work not only validly circumscribed the species, but also noted novel features of its habitat, cyclic migration habits, and cellular biology. This was followed by a study of plastid morphology and development in a number of diverse euglenophytes. The results of this study showed that the plastids of euglenophytes undergo drastic changes in morphology and ultrastructure over the course of a single cell division cycle. I concluded that there are four main classes of plastid development and partitioning in the euglenophytes, and that the class a given species will use is dependant on its interphase plastid morphology and the rigidity of the cell. The discovery of the class IV partitioning strategy in which cells with only one or very few plastids fragment their plastids prior to cell division was very significant. -

Botany Without Bias
editorial Botany without bias In the Gospel According to Matthew Chapter seven, Verse fve, Jesus says “frst cast out the beam out of thine own eye; and then shalt thou see clearly to cast out the mote out of thy brother’s eye”. We should remember this entreaty before too casually casting accusations of ‘plant blindness’. anguage usage helps maintain unconscious biases. In plant biology Lfor example, there is the careless use of the term ‘higher plants’, without thinking about its meaning or implication. If there is a definition of ‘higher plants’ then it is synonymous with vascular plants, but the image it conjures is of upstanding, leafy land-dwelling plants. The problem is that ‘higher’ is a charged term implying superiority of this group over their non-vascular cousins. This stratification is a manifestation of orthogenesis, the idea that evolution has both a direction and a goal. A perfect illustration of orthogenesis is the frequent meme of The Road to Homo Sapiens, the original version of which was drawn by Rudolph Zallinger for a 1965 edition of Life Nature Library1. Also known as The March of Progress, it shows a line of assumed human ancestors, starting with a gibbon-like in their News and Views3, “innovations spend much of their discussions on what Pliopithecus, processing from left to right, associated with improving water use the similarities of these organisms to becoming taller and more upright in stance, efficiency […] may be more fundamental angiosperms can tell about the history and culminating in a modern human. to the evolution of vascular plants than the of plants’ colonization of dry land, and The implication is clear, our evolutionary vascular system from which they derive much less on their characteristics and ancestors are only of interest as waypoints their name”. -

Lateral Gene Transfer of Anion-Conducting Channelrhodopsins Between Green Algae and Giant Viruses
bioRxiv preprint doi: https://doi.org/10.1101/2020.04.15.042127; this version posted April 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 5 Lateral gene transfer of anion-conducting channelrhodopsins between green algae and giant viruses Andrey Rozenberg 1,5, Johannes Oppermann 2,5, Jonas Wietek 2,3, Rodrigo Gaston Fernandez Lahore 2, Ruth-Anne Sandaa 4, Gunnar Bratbak 4, Peter Hegemann 2,6, and Oded 10 Béjà 1,6 1Faculty of Biology, Technion - Israel Institute of Technology, Haifa 32000, Israel. 2Institute for Biology, Experimental Biophysics, Humboldt-Universität zu Berlin, Invalidenstraße 42, Berlin 10115, Germany. 3Present address: Department of Neurobiology, Weizmann 15 Institute of Science, Rehovot 7610001, Israel. 4Department of Biological Sciences, University of Bergen, N-5020 Bergen, Norway. 5These authors contributed equally: Andrey Rozenberg, Johannes Oppermann. 6These authors jointly supervised this work: Peter Hegemann, Oded Béjà. e-mail: [email protected] ; [email protected] 20 ABSTRACT Channelrhodopsins (ChRs) are algal light-gated ion channels widely used as optogenetic tools for manipulating neuronal activity 1,2. Four ChR families are currently known. Green algal 3–5 and cryptophyte 6 cation-conducting ChRs (CCRs), cryptophyte anion-conducting ChRs (ACRs) 7, and the MerMAID ChRs 8. Here we 25 report the discovery of a new family of phylogenetically distinct ChRs encoded by marine giant viruses and acquired from their unicellular green algal prasinophyte hosts. -

Neoproterozoic Origin and Multiple Transitions to Macroscopic Growth in Green Seaweeds
bioRxiv preprint doi: https://doi.org/10.1101/668475; this version posted June 12, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Neoproterozoic origin and multiple transitions to macroscopic growth in green seaweeds Andrea Del Cortonaa,b,c,d,1, Christopher J. Jacksone, François Bucchinib,c, Michiel Van Belb,c, Sofie D’hondta, Pavel Škaloudf, Charles F. Delwicheg, Andrew H. Knollh, John A. Raveni,j,k, Heroen Verbruggene, Klaas Vandepoeleb,c,d,1,2, Olivier De Clercka,1,2 Frederik Leliaerta,l,1,2 aDepartment of Biology, Phycology Research Group, Ghent University, Krijgslaan 281, 9000 Ghent, Belgium bDepartment of Plant Biotechnology and Bioinformatics, Ghent University, Technologiepark 71, 9052 Zwijnaarde, Belgium cVIB Center for Plant Systems Biology, Technologiepark 71, 9052 Zwijnaarde, Belgium dBioinformatics Institute Ghent, Ghent University, Technologiepark 71, 9052 Zwijnaarde, Belgium eSchool of Biosciences, University of Melbourne, Melbourne, Victoria, Australia fDepartment of Botany, Faculty of Science, Charles University, Benátská 2, CZ-12800 Prague 2, Czech Republic gDepartment of Cell Biology and Molecular Genetics, University of Maryland, College Park, MD 20742, USA hDepartment of Organismic and Evolutionary Biology, Harvard University, Cambridge, Massachusetts, 02138, USA. iDivision of Plant Sciences, University of Dundee at the James Hutton Institute, Dundee, DD2 5DA, UK jSchool of Biological Sciences, University of Western Australia (M048), 35 Stirling Highway, WA 6009, Australia kClimate Change Cluster, University of Technology, Ultimo, NSW 2006, Australia lMeise Botanic Garden, Nieuwelaan 38, 1860 Meise, Belgium 1To whom correspondence may be addressed. Email [email protected], [email protected], [email protected] or [email protected]. -

New Phylogenomic Analysis of the Enigmatic Phylum Telonemia Further Resolves the Eukaryote Tree of Life
bioRxiv preprint doi: https://doi.org/10.1101/403329; this version posted August 30, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. New phylogenomic analysis of the enigmatic phylum Telonemia further resolves the eukaryote tree of life Jürgen F. H. Strassert1, Mahwash Jamy1, Alexander P. Mylnikov2, Denis V. Tikhonenkov2, Fabien Burki1,* 1Department of Organismal Biology, Program in Systematic Biology, Uppsala University, Uppsala, Sweden 2Institute for Biology of Inland Waters, Russian Academy of Sciences, Borok, Yaroslavl Region, Russia *Corresponding author: E-mail: [email protected] Keywords: TSAR, Telonemia, phylogenomics, eukaryotes, tree of life, protists bioRxiv preprint doi: https://doi.org/10.1101/403329; this version posted August 30, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Abstract The broad-scale tree of eukaryotes is constantly improving, but the evolutionary origin of several major groups remains unknown. Resolving the phylogenetic position of these ‘orphan’ groups is important, especially those that originated early in evolution, because they represent missing evolutionary links between established groups. Telonemia is one such orphan taxon for which little is known. The group is composed of molecularly diverse biflagellated protists, often prevalent although not abundant in aquatic environments. -
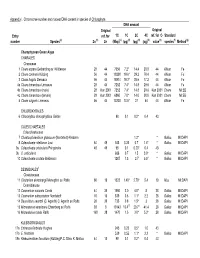
Green Appendix I
Appendix I. Chromsome number and nuclear DNA content in species of Chlorophyta DNA amount Original Original Entry ref. for 1C 1C 2C 4C ref. for C- Standard number Species(a) 2n (b) 2n (Mbp)(c) (pg)(d) (pg)(d) (pg)(d) value(e) species(f) Method(g) Charophycean Green Algae CHARALES Characeae 1 Chara aspera Detharding ex Willdenow 28 44 7056 7.2* 14.4 28.8 44 Allium Fe 2 Chara contraria Kützing 56 44 19208 19.6* 39.2 78.4 44 Allium Fe 3 Chara fragilis Desvaux 56 44 18914 19.3* 38.6 77.2 44 Allium Fe 4a Chara tomentosa Linnaeus 28 44 7252 7.4* 14.8 29.6 44 Allium Fe 4b Chara tomentosa (male) 28 Kun 2001 7252 7.4* 14.8 29.6 Kun 2001 Chara MI:EB 4c Chara tomentsoa (female) 28 Kun 2001 6860 7.0* 14.0 28.0 Kun 2001 Chara MI:EB 5 Chara vulgaris Linnaeus 56 44 13230 13.5* 27 54 44 Allium Fe CHLOROKYBALES 6 Chlorokybus atmosphyticus Geitler 98 0.1 0.2* 0.4 43 COLEOCHAETALES Coleochaetaceae 7 Chaetosphaeridium globosum (Nordstedt) Klebahn 1.2* * Gallus MI:DAPI 8 Coleochaete nitellarum Jost 84 49 343 0.35 0.7 1.4* * Gallus MI:DAPI 9a Coleochaete orbicularis Pringsheim 48 49 98 0.1 0.20* 0.4 43 9b C. orbicularis 686 0.7 1.5 3.0* * Gallus MI:DAPI 10 Coleochaete scutata Brébisson 1287 1.3 2.7 5.5* * Gallus MI:DAPI DESMIDIALES1 Closteriaceae 11 Closterium ehrenbergii Meneghini ex Ralfs 60 19 1323 1.40* 2.70* 5.4 19 Mus MI:DAPI Desmidiaceae 12 Cosmarium cucumis Corda 44 39 1960 2.0 4.0* 8 28 Gallus MI:DAPI 13 Cosmarium subcostatum Nordstedt 10 16 539 0.6 1.1* 2.2 28 Gallus MI:DAPI 14 Desmidium swartzii (C. -

Biosystems Diversity
ISSN 2519-8521 (Print) Regulatory Mechanisms ISSN 2520-2588 (Online) Regul. Mech. Biosyst., 8(4), 532–539 in Biosystems doi: 10.15421/021782 New finding of green algae with potential for algal biotechnology, Chlorococcum oleofaciens and its molecular investigation Y. I. Maltsev, T. V. Konovalenko Bogdan Khmelnitskiy Melitopol State Pedagogical University, Melitopol, Ukraine Article info Maltsev, Y. I., & Konovalenko, T. V. (2017). New finding of green algae with potential for algal biotechnology, Received 14.09.2017 Chlorococcum oleofaciens and its molecular investigation. Regulatory Mechanisms in Biosystems, 8(4), 532–539. Received in revised form doi:10.15421/021782 28.10.2017 Accepted 03.11.2017 The practice of soil algology shows that algae from the order Chlamydomonadales are among the most poorly studied and difficult to identify due to the high heterogeneity of their morphology and ultrastructure. Only the involvement of Bogdan Khmelnitskiy Melitopol molecular genetic methods usually makes it possible to determine their taxonomic status with high accuracy. At the same State Pedagogical University, time, in the algae flora of Ukraine there are more than 250 species from the order Chlamydomonadales, the status of which Getmanska st., 20, Melitopol, in most cases is established exclusively on the basis of light microscopy. This work is devoted to the study of the Zaporizhia region, 72312, Ukraine. biotechnologically promising green alga Chlorococcum oleofaciens, taking into account the modern understanding of its Tel.: +38-096-548-34-36 taxonomic status. Two new strains of this species, separated from samples of forest litter and oak forest soil (the Samara E-mail: [email protected] Forest, Dnipropetrovsk region), are described. -

Different Fates of the Chloroplast Tufa Gene Following Its Transfer to the Nucleus in Green Algae
Proc. Nail. Acad. Sci. USA Vol. 87, pp. 5317-5321, July 1990 Evolution Different fates of the chloroplast tufA gene following its transfer to the nucleus in green algae (elongation factor Tu/gene transfer/organelle genomes/gene duplication/multigene family) SANDRA L. BALDAUF*, JAMES R. MANHARTt, AND JEFFREY D. PALMER* *Department of Biology, Indiana University, Bloomington, IN 47405; and tDepartment of Biology, Texas A&M University, College Station, TX 77843 Communicated by David L. Dilcher, May 8, 1990 (receivedfor review March 23, 1990) ABSTRACT Previous work suggested that the tufA gene, by a combination offilter hybridization and gene sequencing. encoding protein synthesis elongation factor Tu, was trans- Among the Charophyceae, an unusual chloroplast tufAl was ferred from the chloroplast to the nucleus within the green algal found in the genus Coleochaete, the proposed sister group to lineage giving rise to land plants. In this report we investigate land plants (10, 11). the timing and mode of transfer by examining chloroplast and nuclear DNA from the three major classes of green algae, with MATERIALS AND METHODS emphasis on the class Charophyceae, the proposed sister group Filaments of Spirogyra maxima and Sirogonium melanospo- to land plants. Filter hybridizations reveal a chloroplast tufA rum were obtained from unialgal cultures grown in soil/water gene in all Ulvophyceae and Chlorophyceae and in some but not medium (12) on a 16:8 hr light: dark cycle at 200C ± 20C under all Charophyceae. One charophycean alga, Coleochaete orbic- fluorescent light at an illumination of 50 gmol m-2'so1. Ni- ularis, is shown to contain an intact but highly divergent tella translucens and Chara connivens were grown in aquaria chloroplast tufA gene, whose product is predicted to be non- in a soil/water solution in a greenhouse. -

From Algae to Flowering Plants
PP62CH23-Popper ARI 4 April 2011 14:20 Evolution and Diversity of Plant Cell Walls: From Algae to Flowering Plants Zoe¨ A. Popper,1 Gurvan Michel,3,4 Cecile´ Herve,´ 3,4 David S. Domozych,5 William G.T. Willats,6 Maria G. Tuohy,2 Bernard Kloareg,3,4 and Dagmar B. Stengel1 1Botany and Plant Science, and 2Molecular Glycotechnology Group, Biochemistry, School of Natural Sciences, National University of Ireland, Galway, Ireland; email: [email protected] 3CNRS and 4UPMC University Paris 6, UMR 7139 Marine Plants and Biomolecules, Station Biologique de Roscoff, F-29682 Roscoff, Bretagne, France 5Department of Biology and Skidmore Microscopy Imaging Center, Skidmore College, Saratoga Springs, New York 12866 6Department of Plant Biology and Biochemistry, Faculty of Life Sciences, University of Copenhagen, Bulowsvej,¨ 17-1870 Frederiksberg, Denmark Annu. Rev. Plant Biol. 2011. 62:567–90 Keywords First published online as a Review in Advance on xyloglucan, mannan, arabinogalactan proteins, genome, environment, February 22, 2011 multicellularity The Annual Review of Plant Biology is online at plant.annualreviews.org Abstract This article’s doi: All photosynthetic multicellular Eukaryotes, including land plants 10.1146/annurev-arplant-042110-103809 by Universidad Veracruzana on 01/08/14. For personal use only. and algae, have cells that are surrounded by a dynamic, complex, Copyright c 2011 by Annual Reviews. carbohydrate-rich cell wall. The cell wall exerts considerable biologi- All rights reserved cal and biomechanical control over individual cells and organisms, thus Annu. Rev. Plant Biol. 2011.62:567-590. Downloaded from www.annualreviews.org 1543-5008/11/0602-0567$20.00 playing a key role in their environmental interactions. -

The Charophytes (Charophyta) Locality in the Milkha Stream, Lower Jordan, Israel
Natural Resources and Conservation 3(2): 19-30, 2015 http://www.hrpub.org DOI: 10.13189/nrc.2015.030201 The Charophytes (Charophyta) Locality in the Milkha Stream, Lower Jordan, Israel Sophia Barinova1,*, Roman Romanov2 1Institute of Evolution, University of Haifa, Israel 2Central Siberian Botanical Garden of the Siberian Branch of the Russian Academy of Sciences, Russia Copyright © 2015 by authors, all rights reserved. Authors agree that this article remains permanently open access under the terms of the Creative Commons Attribution License 4.0 International License Abstract First study of new locality the Milkha Stream, revealed 14 charophyte species (16 with ifraspecific variety) the Lower Jordan River tributary, with charophyte algae, in that known for Israel [3,4] from references and our studies. semi-arid region of Israel has been implemented for Last year research in Israel and Eastern Mediterranean let us revealing of algal diversity and ecological assessment of the too includes few new localities, algal diversity of which has water object environment by bio-indication methods. been never studied before [5-10]. Altogether forty seven species from five taxonomic The charophytes prefer alkaline water environment which Divisions of algae and cyanobacteria including one of them forms on the carbonates that are very distributed in studied macro-algae Chara gymnophylla A. Braun were revealed in region. This alkaline freshwater environment can be assessed the Milkha stream. Chara was found in growth in the lower as perspective for find new, unstudied aquatic objects in part of studied stream but away from community in followed which can be identified charophyte algae. The most years. -

Table of Contents
Table of Contents General Program………………………………………….. 2 – 5 Poster Presentation Summary……………………………. 6 – 8 Abstracts (in order of presentation)………………………. 9 – 41 Brief Biography, Dr. Dennis Hanisak …………………… 42 1 General Program: 46th Northeast Algal Symposium Friday, April 20, 2007 5:00 – 7:00pm Registration Saturday, April 21, 2007 7:00 – 8:30am Continental Breakfast & Registration 8:30 – 8:45am Welcome and Opening Remarks – Morgan Vis SESSION 1 Student Presentations Moderator: Don Cheney 8:45 – 9:00am Wilce Award Candidate FUSION, DUPLICATION, AND DELETION: EVOLUTION OF EUGLENA GRACILIS LHC POLYPROTEIN-CODING GENES. Adam G. Koziol and Dion G. Durnford. (Abstract p. 9) 9:00 – 9:15am Wilce Award Candidate UTILIZING AN INTEGRATIVE TAXONOMIC APPROACH OF MOLECULAR AND MORPHOLOGICAL CHARACTERS TO DELIMIT SPECIES IN THE RED ALGAL FAMILY KALLYMENIACEAE (RHODOPHYTA). Bridgette Clarkston and Gary W. Saunders. (Abstract p. 9) 9:15 – 9:30am Wilce Award Candidate AFFINITIES OF SOME ANOMALOUS MEMBERS OF THE ACROCHAETIALES. Susan Clayden and Gary W. Saunders. (Abstract p. 10) 9:30 – 9:45am Wilce Award Candidate BARCODING BROWN ALGAE: HOW DNA BARCODING IS CHANGING OUR VIEW OF THE PHAEOPHYCEAE IN CANADA. Daniel McDevit and Gary W. Saunders. (Abstract p. 10) 9:45 – 10:00am Wilce Award Candidate CCMP622 UNID. SP.—A CHLORARACHNIOPHTYE ALGA WITH A ‘LARGE’ NUCLEOMORPH GENOME. Tia D. Silver and John M. Archibald. (Abstract p. 11) 10:00 – 10:15am Wilce Award Candidate PRELIMINARY INVESTIGATION OF THE NUCLEOMORPH GENOME OF THE SECONDARILY NON-PHOTOSYNTHETIC CRYPTOMONAD CRYPTOMONAS PARAMECIUM CCAP977/2A. Natalie Donaher, Christopher Lane and John Archibald. (Abstract p. 11) 10:15 – 10:45am Break 2 SESSION 2 Student Presentations Moderator: Hilary McManus 10:45 – 11:00am Wilce Award Candidate IMPACTS OF HABITAT-MODIFYING INVASIVE MACROALGAE ON EPIPHYTIC ALGAL COMMUNTIES.