Biosystems Diversity
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Chlorophyta, Trebouxiophyceae) in Lake Tanganyika (Africa)*
Biologia 63/6: 799—805, 2008 Section Botany DOI: 10.2478/s11756-008-0101-4 Siderocelis irregularis (Chlorophyta, Trebouxiophyceae) in Lake Tanganyika (Africa)* Maya P. Stoyneva1, Elisabeth Ingolič2,WernerKofler3 &WimVyverman4 1Sofia University ‘St Kliment Ohridski’, Faculty of Biology, Department of Botany, 8 bld. Dragan Zankov, BG-1164 Sofia, Bulgaria; e-mail: [email protected], [email protected]fia.bg 2Graz University of Technology, Research Institute for Electron Microscopy, Steyrergasse 17,A-8010 Graz, Austria; e-mail: [email protected] 3University of Innsbruck, Institute of Botany, Sternwartestrasse 15,A-6020 Innsbruck, Austria; e-mail: werner.kofl[email protected] 4Ghent University, Department Biology, Laboratory of Protistology and Aquatic Ecology, Krijgslaan 281-S8,B-9000 Gent, Belgium; e-mail: [email protected] Abstract: Siderocelis irregularis Hindák, representing a genus Siderocelis (Naumann) Fott that is known from European temperate waters, was identified as a common phytoplankter in Lake Tanganyika. It was found aposymbiotic as well as ingested (possibly endosymbiotic) in lake heterotrophs, mainly Strombidium sp. and Vorticella spp. The morphology and ultrastructure of the species, studied with LM, SEM and TEM, are described with emphasis on the structure of the cell wall and the pyrenoid. Key words: Chlorophyta; cell wall; pyrenoid; symbiosis; ciliates; Strombidium; Vorticella Introduction ics of symbiotic species in general came into alignment with that of free-living algae and the term ‘zoochlorel- Tight partnerships between algae and aquatic inver- lae’ was abandoned as being taxonomically ambiguous tebrates, including symbiotic relationships, have long (e.g. Bal 1968; Reisser & Wiessner 1984; Taylor 1984; been of interest and a number of excellent reviews are Reisser 1992a). -

Neoproterozoic Origin and Multiple Transitions to Macroscopic Growth in Green Seaweeds
Neoproterozoic origin and multiple transitions to macroscopic growth in green seaweeds Andrea Del Cortonaa,b,c,d,1, Christopher J. Jacksone, François Bucchinib,c, Michiel Van Belb,c, Sofie D’hondta, f g h i,j,k e Pavel Skaloud , Charles F. Delwiche , Andrew H. Knoll , John A. Raven , Heroen Verbruggen , Klaas Vandepoeleb,c,d,1,2, Olivier De Clercka,1,2, and Frederik Leliaerta,l,1,2 aDepartment of Biology, Phycology Research Group, Ghent University, 9000 Ghent, Belgium; bDepartment of Plant Biotechnology and Bioinformatics, Ghent University, 9052 Zwijnaarde, Belgium; cVlaams Instituut voor Biotechnologie Center for Plant Systems Biology, 9052 Zwijnaarde, Belgium; dBioinformatics Institute Ghent, Ghent University, 9052 Zwijnaarde, Belgium; eSchool of Biosciences, University of Melbourne, Melbourne, VIC 3010, Australia; fDepartment of Botany, Faculty of Science, Charles University, CZ-12800 Prague 2, Czech Republic; gDepartment of Cell Biology and Molecular Genetics, University of Maryland, College Park, MD 20742; hDepartment of Organismic and Evolutionary Biology, Harvard University, Cambridge, MA 02138; iDivision of Plant Sciences, University of Dundee at the James Hutton Institute, Dundee DD2 5DA, United Kingdom; jSchool of Biological Sciences, University of Western Australia, WA 6009, Australia; kClimate Change Cluster, University of Technology, Ultimo, NSW 2006, Australia; and lMeise Botanic Garden, 1860 Meise, Belgium Edited by Pamela S. Soltis, University of Florida, Gainesville, FL, and approved December 13, 2019 (received for review June 11, 2019) The Neoproterozoic Era records the transition from a largely clear interpretation of how many times and when green seaweeds bacterial to a predominantly eukaryotic phototrophic world, creat- emerged from unicellular ancestors (8). ing the foundation for the complex benthic ecosystems that have There is general consensus that an early split in the evolution sustained Metazoa from the Ediacaran Period onward. -

JUDD W.S. Et. Al. (2002) Plant Systematics: a Phylogenetic Approach. Chapter 7. an Overview of Green
UNCORRECTED PAGE PROOFS An Overview of Green Plant Phylogeny he word plant is commonly used to refer to any auto- trophic eukaryotic organism capable of converting light energy into chemical energy via the process of photosynthe- sis. More specifically, these organisms produce carbohydrates from carbon dioxide and water in the presence of chlorophyll inside of organelles called chloroplasts. Sometimes the term plant is extended to include autotrophic prokaryotic forms, especially the (eu)bacterial lineage known as the cyanobacteria (or blue- green algae). Many traditional botany textbooks even include the fungi, which differ dramatically in being heterotrophic eukaryotic organisms that enzymatically break down living or dead organic material and then absorb the simpler products. Fungi appear to be more closely related to animals, another lineage of heterotrophs characterized by eating other organisms and digesting them inter- nally. In this chapter we first briefly discuss the origin and evolution of several separately evolved plant lineages, both to acquaint you with these important branches of the tree of life and to help put the green plant lineage in broad phylogenetic perspective. We then focus attention on the evolution of green plants, emphasizing sev- eral critical transitions. Specifically, we concentrate on the origins of land plants (embryophytes), of vascular plants (tracheophytes), of 1 UNCORRECTED PAGE PROOFS 2 CHAPTER SEVEN seed plants (spermatophytes), and of flowering plants dons.” In some cases it is possible to abandon such (angiosperms). names entirely, but in others it is tempting to retain Although knowledge of fossil plants is critical to a them, either as common names for certain forms of orga- deep understanding of each of these shifts and some key nization (e.g., the “bryophytic” life cycle), or to refer to a fossils are mentioned, much of our discussion focuses on clade (e.g., applying “gymnosperms” to a hypothesized extant groups. -

The Green Puzzle Stichococcus (Trebouxiophyceae, Chlorophyta): New Generic and Species Concept Among This Widely Distributed Genus
Phytotaxa 441 (2): 113–142 ISSN 1179-3155 (print edition) https://www.mapress.com/j/pt/ PHYTOTAXA Copyright © 2020 Magnolia Press Article ISSN 1179-3163 (online edition) https://doi.org/10.11646/phytotaxa.441.2.2 The green puzzle Stichococcus (Trebouxiophyceae, Chlorophyta): New generic and species concept among this widely distributed genus THOMAS PRÖSCHOLD1,3* & TATYANA DARIENKO2,4 1 University of Innsbruck, Research Department for Limnology, A-5310 Mondsee, Austria 2 University of Göttingen, Albrecht-von-Haller-Institute of Plant Sciences, Experimental Phycology and Sammlung für Algenkulturen, D-37073 Göttingen, Germany 3 [email protected]; http://orcid.org/0000-0002-7858-0434 4 [email protected]; http://orcid.org/0000-0002-1957-0076 *Correspondence author Abstract Phylogenetic analyses have revealed that the traditional order Prasiolales, which contains filamentous and pseudoparenchy- matous genera Prasiola and Rosenvingiella with complex life cycle, also contains taxa of more simple morphology such as coccoids like Pseudochlorella and Edaphochlorella or rod-like organisms like Stichococcus and Pseudostichococcus (called Prasiola clade of the Trebouxiophyceae). Recent studies have shown a high biodiversity among these organisms and questioned the traditional generic and species concept. We studied 34 strains assigned as Stichococcus, Pseudostichococcus, Diplosphaera and Desmocococcus. Phylogenetic analyses using a multigene approach revealed that these strains belong to eight independent lineages within the Prasiola clade of the Trebouxiophyceae. For testing if these lineages represent genera, we studied the secondary structures of SSU and ITS rDNA sequences to find genetic synapomorphies. The secondary struc- ture of the V9 region of SSU is diagnostic to support the proposal for separation of eight genera. -
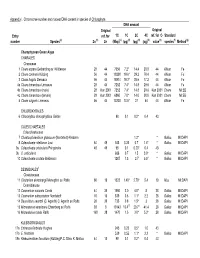
Green Appendix I
Appendix I. Chromsome number and nuclear DNA content in species of Chlorophyta DNA amount Original Original Entry ref. for 1C 1C 2C 4C ref. for C- Standard number Species(a) 2n (b) 2n (Mbp)(c) (pg)(d) (pg)(d) (pg)(d) value(e) species(f) Method(g) Charophycean Green Algae CHARALES Characeae 1 Chara aspera Detharding ex Willdenow 28 44 7056 7.2* 14.4 28.8 44 Allium Fe 2 Chara contraria Kützing 56 44 19208 19.6* 39.2 78.4 44 Allium Fe 3 Chara fragilis Desvaux 56 44 18914 19.3* 38.6 77.2 44 Allium Fe 4a Chara tomentosa Linnaeus 28 44 7252 7.4* 14.8 29.6 44 Allium Fe 4b Chara tomentosa (male) 28 Kun 2001 7252 7.4* 14.8 29.6 Kun 2001 Chara MI:EB 4c Chara tomentsoa (female) 28 Kun 2001 6860 7.0* 14.0 28.0 Kun 2001 Chara MI:EB 5 Chara vulgaris Linnaeus 56 44 13230 13.5* 27 54 44 Allium Fe CHLOROKYBALES 6 Chlorokybus atmosphyticus Geitler 98 0.1 0.2* 0.4 43 COLEOCHAETALES Coleochaetaceae 7 Chaetosphaeridium globosum (Nordstedt) Klebahn 1.2* * Gallus MI:DAPI 8 Coleochaete nitellarum Jost 84 49 343 0.35 0.7 1.4* * Gallus MI:DAPI 9a Coleochaete orbicularis Pringsheim 48 49 98 0.1 0.20* 0.4 43 9b C. orbicularis 686 0.7 1.5 3.0* * Gallus MI:DAPI 10 Coleochaete scutata Brébisson 1287 1.3 2.7 5.5* * Gallus MI:DAPI DESMIDIALES1 Closteriaceae 11 Closterium ehrenbergii Meneghini ex Ralfs 60 19 1323 1.40* 2.70* 5.4 19 Mus MI:DAPI Desmidiaceae 12 Cosmarium cucumis Corda 44 39 1960 2.0 4.0* 8 28 Gallus MI:DAPI 13 Cosmarium subcostatum Nordstedt 10 16 539 0.6 1.1* 2.2 28 Gallus MI:DAPI 14 Desmidium swartzii (C. -

Plastid Genomes of the Early Vascular Plant Genus Selaginella Have Unusual Direct Repeat Structures and Drastically Reduced Gene Numbers
International Journal of Molecular Sciences Article Plastid Genomes of the Early Vascular Plant Genus Selaginella Have Unusual Direct Repeat Structures and Drastically Reduced Gene Numbers Hyeonah Shim 1, Hyeon Ju Lee 1, Junki Lee 1,2, Hyun-Oh Lee 1,2, Jong-Hwa Kim 3, Tae-Jin Yang 1,* and Nam-Soo Kim 4,* 1 Department of Agriculture, Forestry and Bioresources, Plant Genomics & Breeding Institute, Research Institute of Agriculture and Life Sciences, College of Agriculture & Life Sciences, Seoul National University, 1 Gwanak-ro, Gwanak-gu, Seoul 08826, Korea; [email protected] (H.S.); [email protected] (H.J.L.); [email protected] (J.L.); [email protected] (H.-O.L.) 2 Phyzen Genomics Institute, Seongnam 13558, Korea 3 Department of Horticulture, Kangwon National University, Chuncheon 24341, Korea; [email protected] 4 Department of Molecular Bioscience, Kangwon National University, Chuncheon 24341, Korea * Correspondence: [email protected] (T.-J.Y.); [email protected] (N.-S.K.); Tel.: +82-2-880-4547 (T.-J.Y.); +82-33-250-6472 (N.-S.K.) Abstract: The early vascular plants in the genus Selaginella, which is the sole genus of the Selaginel- laceae family, have an important place in evolutionary history, along with ferns, as such plants are valuable resources for deciphering plant evolution. In this study, we sequenced and assembled the plastid genome (plastome) sequences of two Selaginella tamariscina individuals, as well as Se- laginella stauntoniana and Selaginella involvens. Unlike the inverted repeat (IR) structures typically found in plant plastomes, Selaginella species had direct repeat (DR) structures, which were confirmed by Oxford Nanopore long-read sequence assembly. -

From Algae to Flowering Plants
PP62CH23-Popper ARI 4 April 2011 14:20 Evolution and Diversity of Plant Cell Walls: From Algae to Flowering Plants Zoe¨ A. Popper,1 Gurvan Michel,3,4 Cecile´ Herve,´ 3,4 David S. Domozych,5 William G.T. Willats,6 Maria G. Tuohy,2 Bernard Kloareg,3,4 and Dagmar B. Stengel1 1Botany and Plant Science, and 2Molecular Glycotechnology Group, Biochemistry, School of Natural Sciences, National University of Ireland, Galway, Ireland; email: [email protected] 3CNRS and 4UPMC University Paris 6, UMR 7139 Marine Plants and Biomolecules, Station Biologique de Roscoff, F-29682 Roscoff, Bretagne, France 5Department of Biology and Skidmore Microscopy Imaging Center, Skidmore College, Saratoga Springs, New York 12866 6Department of Plant Biology and Biochemistry, Faculty of Life Sciences, University of Copenhagen, Bulowsvej,¨ 17-1870 Frederiksberg, Denmark Annu. Rev. Plant Biol. 2011. 62:567–90 Keywords First published online as a Review in Advance on xyloglucan, mannan, arabinogalactan proteins, genome, environment, February 22, 2011 multicellularity The Annual Review of Plant Biology is online at plant.annualreviews.org Abstract This article’s doi: All photosynthetic multicellular Eukaryotes, including land plants 10.1146/annurev-arplant-042110-103809 by Universidad Veracruzana on 01/08/14. For personal use only. and algae, have cells that are surrounded by a dynamic, complex, Copyright c 2011 by Annual Reviews. carbohydrate-rich cell wall. The cell wall exerts considerable biologi- All rights reserved cal and biomechanical control over individual cells and organisms, thus Annu. Rev. Plant Biol. 2011.62:567-590. Downloaded from www.annualreviews.org 1543-5008/11/0602-0567$20.00 playing a key role in their environmental interactions. -

Compartmentalization of Mrnas in the Giant, Unicellular Green Algae
bioRxiv preprint doi: https://doi.org/10.1101/2020.09.18.303206; this version posted September 18, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Compartmentalization of mRNAs in the giant, 2 unicellular green algae Acetabularia acetabulum 3 4 Authors 5 Ina J. Andresen1, Russell J. S. Orr2, Kamran Shalchian-Tabrizi3, Jon Bråte1* 6 7 Address 8 1: Section for Genetics and Evolutionary Biology, Department of Biosciences, University of 9 Oslo, Kristine Bonnevies Hus, Blindernveien 31, 0316 Oslo, Norway. 10 2: Natural History Museum, University of Oslo, Oslo, Norway 11 3: Centre for Epigenetics, Development and Evolution, Department of Biosciences, University 12 of Oslo, Kristine Bonnevies Hus, Blindernveien 31, 0316 Oslo, Norway. 13 14 *Corresponding author 15 Jon Bråte, [email protected] 16 17 Keywords 18 Acetabularia acetabulum, Dasycladales, UMI, STL, compartmentalization, single-cell, mRNA. 19 20 Abstract 21 Acetabularia acetabulum is a single-celled green alga previously used as a model species for 22 studying the role of the nucleus in cell development and morphogenesis. The highly elongated 23 cell, which stretches several centimeters, harbors a single nucleus located in the basal end. 24 Although A. acetabulum historically has been an important model in cell biology, almost 25 nothing is known about its gene content, or how gene products are distributed in the cell. To 26 study the composition and distribution of mRNAs in A. -

The Charophytes (Charophyta) Locality in the Milkha Stream, Lower Jordan, Israel
Natural Resources and Conservation 3(2): 19-30, 2015 http://www.hrpub.org DOI: 10.13189/nrc.2015.030201 The Charophytes (Charophyta) Locality in the Milkha Stream, Lower Jordan, Israel Sophia Barinova1,*, Roman Romanov2 1Institute of Evolution, University of Haifa, Israel 2Central Siberian Botanical Garden of the Siberian Branch of the Russian Academy of Sciences, Russia Copyright © 2015 by authors, all rights reserved. Authors agree that this article remains permanently open access under the terms of the Creative Commons Attribution License 4.0 International License Abstract First study of new locality the Milkha Stream, revealed 14 charophyte species (16 with ifraspecific variety) the Lower Jordan River tributary, with charophyte algae, in that known for Israel [3,4] from references and our studies. semi-arid region of Israel has been implemented for Last year research in Israel and Eastern Mediterranean let us revealing of algal diversity and ecological assessment of the too includes few new localities, algal diversity of which has water object environment by bio-indication methods. been never studied before [5-10]. Altogether forty seven species from five taxonomic The charophytes prefer alkaline water environment which Divisions of algae and cyanobacteria including one of them forms on the carbonates that are very distributed in studied macro-algae Chara gymnophylla A. Braun were revealed in region. This alkaline freshwater environment can be assessed the Milkha stream. Chara was found in growth in the lower as perspective for find new, unstudied aquatic objects in part of studied stream but away from community in followed which can be identified charophyte algae. The most years. -

Seaweeds of California Green Algae
PDF version Remove references Seaweeds of California (draft: Sun Nov 24 15:32:39 2019) This page provides current names for California seaweed species, including those whose names have changed since the publication of Marine Algae of California (Abbott & Hollenberg 1976). Both former names (1976) and current names are provided. This list is organized by group (green, brown, red algae); within each group are genera and species in alphabetical order. California seaweeds discovered or described since 1976 are indicated by an asterisk. This is a draft of an on-going project. If you have questions or comments, please contact Kathy Ann Miller, University Herbarium, University of California at Berkeley. [email protected] Green Algae Blidingia minima (Nägeli ex Kützing) Kylin Blidingia minima var. vexata (Setchell & N.L. Gardner) J.N. Norris Former name: Blidingia minima var. subsalsa (Kjellman) R.F. Scagel Current name: Blidingia subsalsa (Kjellman) R.F. Scagel et al. Kornmann, P. & Sahling, P.H. 1978. Die Blidingia-Arten von Helgoland (Ulvales, Chlorophyta). Helgoländer Wissenschaftliche Meeresuntersuchungen 31: 391-413. Scagel, R.F., Gabrielson, P.W., Garbary, D.J., Golden, L., Hawkes, M.W., Lindstrom, S.C., Oliveira, J.C. & Widdowson, T.B. 1989. A synopsis of the benthic marine algae of British Columbia, southeast Alaska, Washington and Oregon. Phycological Contributions, University of British Columbia 3: vi + 532. Bolbocoleon piliferum Pringsheim Bryopsis corticulans Setchell Bryopsis hypnoides Lamouroux Former name: Bryopsis pennatula J. Agardh Current name: Bryopsis pennata var. minor J. Agardh Silva, P.C., Basson, P.W. & Moe, R.L. 1996. Catalogue of the benthic marine algae of the Indian Ocean. -

Unraveling the Photoprotective Response of Lichenized and Free-Living Green Algae (Trebouxiophyceae, Chlorophyta) to Photochilling Stress
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Archivo Digital para la Docencia y la Investigación ORIGINAL RESEARCH published: 04 July 2017 doi: 10.3389/fpls.2017.01144 Unraveling the Photoprotective Response of Lichenized and Free-Living Green Algae (Trebouxiophyceae, Chlorophyta) to Photochilling Stress Fátima Míguez 1*, Ulf Schiefelbein 2, Ulf Karsten 3, José I. García-Plazaola 1 and Lydia Gustavs 3 1 Department of Plant Biology and Ecology, University of the Basque Country (UPV/EHU), Bilbao, Spain, 2 Independant Researcher, Rostock, Germany, 3 Applied Ecology and Phycology, Institute of Biological Sciences, University of Rostock, Rostock, Germany Lichens and free-living terrestrial algae are widespread across many habitats and develop successfully in ecosystems where a cold winter limits survival. With the Edited by: goal of comparing photoprotective responses in free-living and lichenized algae, the Tiina Tosens, physiological responses to chilling and photochilling conditions were studied in three Estonian University of Life Sciences, lichens and their isolated algal photobionts together as well as in a fourth free-living Estonia algal species. We specifically addressed the following questions: (i) Are there general Reviewed by: Pille Mänd, patterns of acclimation in green algae under chilling and photochilling stresses? (ii) Do University of Tartu, Estonia free-living algae exhibit a similar pattern of responses as their lichenized counterparts? (iii) Lea Hallik, Tartu Observatory (ETA), Estonia Are these responses influenced by the selection pressure of environmental conditions or *Correspondence: by the phylogenetic position of each species? To answer these questions, photosynthetic Fátima Míguez fluorescence measurements as well as pigment and low molecular weight carbohydrate [email protected] pool analyses were performed under controlled laboratory conditions. -

The Marine Vegetation of the Kerguelen Islands: History of Scientific Campaigns, Inventory of the Flora and First Analysis of Its Biogeographical Affinities
cryptogamie Algologie 2021 ● 42 ● 12 DIRECTEUR DE LA PUBLICATION / PUBLICATION DIRECTOR : Bruno DAVID Président du Muséum national d’Histoire naturelle RÉDACTRICE EN CHEF / EDITOR-IN-CHIEF : Line LE GALL Muséum national d’Histoire naturelle ASSISTANTE DE RÉDACTION / ASSISTANT EDITOR : Marianne SALAÜN ([email protected]) MISE EN PAGE / PAGE LAYOUT : Marianne SALAÜN RÉDACTEURS ASSOCIÉS / ASSOCIATE EDITORS Ecoevolutionary dynamics of algae in a changing world Stacy KRUEGER-HADFIELD Department of Biology, University of Alabama, 1300 University Blvd, Birmingham, AL 35294 (United States) Jana KULICHOVA Department of Botany, Charles University, Prague (Czech Republic) Cecilia TOTTI Dipartimento di Scienze della Vita e dell’Ambiente, Università Politecnica delle Marche, Via Brecce Bianche, 60131 Ancona (Italy) Phylogenetic systematics, species delimitation & genetics of speciation Sylvain FAUGERON UMI3614 Evolutionary Biology and Ecology of Algae, Departamento de Ecología, Facultad de Ciencias Biologicas, Pontificia Universidad Catolica de Chile, Av. Bernardo O’Higgins 340, Santiago (Chile) Marie-Laure GUILLEMIN Instituto de Ciencias Ambientales y Evolutivas, Universidad Austral de Chile, Valdivia (Chile) Diana SARNO Department of Integrative Marine Ecology, Stazione Zoologica Anton Dohrn, Villa Comunale, 80121 Napoli (Italy) Comparative evolutionary genomics of algae Nicolas BLOUIN Department of Molecular Biology, University of Wyoming, Dept. 3944, 1000 E University Ave, Laramie, WY 82071 (United States) Heroen VERBRUGGEN School of BioSciences,