A Remote Response of ATP Hydrolysis Xin Liu State Key Laboratory of Nonlinear Mechanics
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

AP Biology Names___Biomolecule Stations Per
AP Biology Names____________________________________ Biomolecule Stations Per.______________ In this two-day activity, you will move through several different stations and learn about the four macromolecules in the biological world. Day 1: Modeling Carbohydrates and Lipids 1. Use the 3d-printed models to answer the questions for carbohydrates and lipids. NOTE: Blank corners indicate “carbon” atoms. Glucose Fructose Glycerol Acetic Acid Butyric Acid Caproic Acid Day 2: Modeling Proteins 1. Using the Amino Acid Sidechain list, organize the sidechains on the circular magnetic mat according to their name or properties. 2. Examine the side chains and their positions on the circle. Describe your observations on your answer sheet by answering questions #1-6 under Modeling Proteins. 3. Now you have explored the chemical properties and atomic composition of each sidechain, you are ready to predict how proteins spontaneously fold up into their 3D shapes. Answer questions #7-8 on your answer sheet. 4. Unwind the yellow tube. Notice the blue and red end caps. The blue end cap represents the N- terminus (the beginning) and the red end cap represents the C-terminus (the end) of the protein. 5. Select 15 metal U-shaped clips from your kit. Beginning at the N-terminus, place the 15 u-clips three inches apart on the tube. 6. Select methionine from the mat and place it on the clip closest to the blue end cap. Choose six hydrophobic sidechains, two acidic sidechains, two basic sidechains, two cysteine sidechains, and two other polar sidechains. Place them in any order you choose on your tubes u-clips. -

Biology (BIOL) 1
Biology (BIOL) 1 Biology (BIOL) Courses BIOL 0848. DNA: Friend or Foe. 3 Credit Hours. This course is typically offered in Fall. Through the study of basic biological concepts, think critically about modern biotechnology. Consider questions like: What are the ethical and legal implications involving the gathering and analysis of DNA samples for forensic analysis and DNA fingerprinting? Are there potential discriminatory implications that might result from the human genome project? What are embryonic stem cells, and why has this topic become an important social and political issue? Will advances in medicine allow humans to live considerably longer, and how will a longer human life span affect life on earth? We will learn through lectures, lecture demonstrations, problem solving in small groups and classroom discussion, and make vivid use of technology, including short videos from internet sources such as YouTube, electronic quizzes, imaging and video microscopy. NOTE: This course fulfills a Science & Technology (GS) requirement for students under GenEd and the Science & Technology Second Level (SB) requirement for students under Core. Students cannot receive credit for this course if they have successfully completed Biology 0948. Course Attributes: GS Repeatability: This course may not be repeated for additional credits. BIOL 0948. Honors DNA: Friend or Foe. 3 Credit Hours. This course is not offered every year. Through the study of basic biological concepts, think critically about modern biotechnology. Consider questions like: What are -

A Resonant Capacitive Test Structure for Biomolecule Sensing
A RESONANT CAPACITIVE TEST STRUCTURE FOR BIOMOLECULE SENSING Thesis Submitted to The School of Engineering of the UNIVERSITY OF DAYTON In Partial Fulfillment of the Requirements for The Degree of Master of Science in Electrical Engineering By Danielle Nichole Bane UNIVERSITY OF DAYTON Dayton, Ohio August, 2015 A RESONANT CAPACITIVE TEST STRUCTURE FOR BIOMOLECULE SENSING Name: Bane, Danielle Nichole APPROVED BY: Guru Subramanyam, Ph.D. Karolyn M. Hansen, Ph.D. Advisor Committee Chairman Advisor Committee Member Professor and Chair, Department of Professor, Department of Biology Electrical and Computer Engineering Partha Banerjee, Ph.D Committee Member Professor, Department of Electrical and Computer Engineering John G. Weber, Ph.D. Eddy M. Rojas, Ph.D., M.A., P.E. Associate Dean Dean School of Engineering School of Engineering ii c Copyright by Danielle Nichole Bane All rights reserved 2015 ABSTRACT A RESONANT CAPACITIVE TEST STRUCTURE FOR BIOMOLECULE SENSING Name: Bane, Danielle Nichole University of Dayton Advisors: Dr. Guru Subramanyam and Dr. Karolyn M. Hansen Detection of biomolecules in aqueous or vapor phase is a valuable metric in the assessment of health and human performance. For this purpose, resonant capacitive sensors are designed and fab- ricated. The sensor platform used is a resonant test structure (RTS) with a molecular recognition element (MRE) functionalized guanine dielectric layer used as the sensing layer. The sensors are designed such that the selective binding of the biomarkers of interest with the MREs is expected to cause a shift in the test structure’s resonant frequency, amplitude, and phase thereby indicating the biomarker’s presence. This thesis covers several aspects of the design and development of these biosensors. -

Metabolic Fate of Glucose Metabolic Fate of Fatty Acids
CHEM464/Medh, J.D. Integration of Metabolism Metabolic Fate of Glucose • Each class of biomolecule has alternative fates depending on the metabolic state of the body. • Glucose: The intracellular form of glucose is glucose-6- phosphate. • Only liver cells have the enzyme glucose-6-phosphatase that dephosphorylates G-6-P and releases glucose into the blood for use by other tissues • G-6-P can be oxidized for energy in the form of ATP and NADH • G-6-P can be converted to acetyl CoA and then fat. • Excess G-6-P is stored away as glycogen. • G-6-P can be shunted into the pentose phosphate pathway to generate NADPH and ribose-5-phosphate. Metabolic Fate of Fatty Acids • Fatty acids are oxidized to acetyl CoA for energy production in the form of NADH. • Fatty acids can be converted to ketone bodies. KB can be used as fuel in extrahepatic tissues. • Palmityl CoA is a precursor of mono- and poly- unsaturated fatty acids. • Fatty acids are used for the biosynthesis of bioactive molecules such as arachidonic acid and eicosanoids. • Cholesterol, steroids and steroid hormones are all derived from fatty acids. • Excess fatty acids are stored away as triglycerides in adipose tissue. 1 CHEM464/Medh, J.D. Integration of Metabolism Metabolic Fate of Amino Acids • Amino acids are used for the synthesis of enzymes, transporters and other physiologically significant proteins. • Amino acid N is required for synthesis of the cell’s genetic information (synthesis of nitrogenous bases). • Several biologically active molecules such as neuro- transmitters, porphyrins etc. • Amino acids are precursors of several hormones (peptide hormones like insulin and glucagon and Amine hormones such as catecholamines). -

Biomolecule and Bioentity Interaction Databases in Systems Biology: a Comprehensive Review
biomolecules Review Biomolecule and Bioentity Interaction Databases in Systems Biology: A Comprehensive Review Fotis A. Baltoumas 1,* , Sofia Zafeiropoulou 1, Evangelos Karatzas 1 , Mikaela Koutrouli 1,2, Foteini Thanati 1, Kleanthi Voutsadaki 1 , Maria Gkonta 1, Joana Hotova 1, Ioannis Kasionis 1, Pantelis Hatzis 1,3 and Georgios A. Pavlopoulos 1,3,* 1 Institute for Fundamental Biomedical Research, Biomedical Sciences Research Center “Alexander Fleming”, 16672 Vari, Greece; zafeiropoulou@fleming.gr (S.Z.); karatzas@fleming.gr (E.K.); [email protected] (M.K.); [email protected] (F.T.); voutsadaki@fleming.gr (K.V.); [email protected] (M.G.); hotova@fleming.gr (J.H.); [email protected] (I.K.); hatzis@fleming.gr (P.H.) 2 Novo Nordisk Foundation Center for Protein Research, University of Copenhagen, 2200 Copenhagen, Denmark 3 Center for New Biotechnologies and Precision Medicine, School of Medicine, National and Kapodistrian University of Athens, 11527 Athens, Greece * Correspondence: baltoumas@fleming.gr (F.A.B.); pavlopoulos@fleming.gr (G.A.P.); Tel.: +30-210-965-6310 (G.A.P.) Abstract: Technological advances in high-throughput techniques have resulted in tremendous growth Citation: Baltoumas, F.A.; of complex biological datasets providing evidence regarding various biomolecular interactions. Zafeiropoulou, S.; Karatzas, E.; To cope with this data flood, computational approaches, web services, and databases have been Koutrouli, M.; Thanati, F.; Voutsadaki, implemented to deal with issues such as data integration, visualization, exploration, organization, K.; Gkonta, M.; Hotova, J.; Kasionis, scalability, and complexity. Nevertheless, as the number of such sets increases, it is becoming more I.; Hatzis, P.; et al. Biomolecule and and more difficult for an end user to know what the scope and focus of each repository is and how Bioentity Interaction Databases in redundant the information between them is. -

Biomolecules Review
Biomolecules Review Read the description. Hold up the card for the correct biomolecule when I ask you to do so. Which biomolecule is this? • CARBOHYDRATE Which biomolecule is this? • Protein Which biomolecule is this? • Lipid Which biomolecule is this? • Carbohydrate Which biomolecule is this? • Protein Which biomolecule is this? • Nucleic Acid Which biomolecule is this? • Carbohydrate Which biomolecule is this? • Lipid Which biomolecule is this? • Nucleic Acid Which biomolecule is this? • Protein Which biomolecule is this? • Nucleic Acid Which biomolecule is this? • Nucleic Acid Which biomolecule is this? • Proteins Which biomolecule is this? • Lipid Which biomolecule is this? • Carbohydrate Which biomolecule is this? • Protein Which biomolecule is this? • Lipids Which biomolecule is this? • Nucleic Acid Which biomolecule is this? • Carbohydrate Which biomolecule has this function? • Stores and transmits heredity or genetic information • Nucleic Acid Which biomolecule has this function? • Cellulose provides support and structure for plants. • Carbohydrates Which biomolecule has this function? • Antibodies help defend against disease and fight infections. • Proteins Which biomolecule has this function? • An important part of cell membranes • Lipids Which biomolecule has this function? • Transports substances in and out of cells through channels in the cell membrane. • Proteins Which biomolecule has this function? • Used to store energy that is released slowly. • Lipids Which biomolecule has this function? • Forms bones, muscles, hair and nails. • Proteins Which biomolecule has this function? • Main source of quick energy for living things. • Carbohydrates Which biomolecule has this function? • Helps to regulate cell processes by the use of hormones. • Proteins Which biomolecule has this function? • Starch is the main form of stored energy for plants. -

Biomolecular Materials. Report of the January 13-15, 2002 Workshop
Cover Illustrations (from top): Molecular graphics representation looking into the channel of the a hemolysin pore. Song!et al., 1996 (Figure 24). Complexation of F-actin and cationic lipids leads to the hierarchical self-assembly of a network of tubules; shown here in cross-section. Wong 2000 (Figure 9). Depiction of an array of hybrid nanodevices powered by F1-ATPase. Soong et al., 2000 (Figure 1). Electronmicrograph, after fixation, of neuron from the A cluster of the pedal ganglia in L. stagnalis immobilized within a picket fence of polyimide after 3 days in culture on silicon chip. (Scale bar = 20!mm.). Zeck and Fromherz 2001 (Figure 16). Biomolecular Materials Report of the January 13-15, 2002 Workshop Supported by the Basic Energy Sciences Advisory Committee U.S. Department of Energy Co-Chairs: Mark D. Alper Materials Sciences Division Lawrence Berkeley National Laboratory and Department of Molecular and Cell Biology University of California at Berkeley Berkeley, CA 94720 Samuel I. Stupp Department of Materials Science and Engineering Department of Chemistry Feinberg School of Medicine Northwestern University Evanston, IL 60208 This work was supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under contract DE-AC03-76SF00098 This report is dedicated to the memory of Iran L. Thomas, Deputy Director of the Department of Energy Office of Basic Energy Sciences and Director of the Division of Materials Sciences and Engineering until his death in February, 2003. Iran was among the first to recognize the potential impact of the biological sciences on the physical sciences, organizing workshops and initiating programs in that area as early as 1988. -

Biomolecule Activity AP Biology - Biomolecule Activity
AP Biology - Biomolecule Activity AP Biology - Biomolecule Activity Perform the following tasks and write your responses in your lab notebook. Perform the following tasks and write your responses in your lab notebook. Carbohydrates Carbohydrates 1. Each team of 2 makes a glucose molecule or a fructose molecule. Then through a 1. Each team of 2 makes a glucose molecule or a fructose molecule. Then through a condensation reaction, synthesize one model of the disaccharide sucrose. See condensation reaction, synthesize one model of the disaccharide sucrose. See Section Section 2.3 and Figure 2.9 in PoL textbook. 2.3 and Figure 2.9 in PoL textbook. 2. Show Ms. Morgan your fructose and water molecules. 2. Show Ms. Morgan your fructose and water molecules. 3. Using PoL or other resources, locate and write down the following: 3. Using PoL or other resources, locate and write down the following: a. The names and biological functions of 3 monosaccharides, 3 a. The names and biological functions of 3 monosaccharides, 3 disaccharides, disaccharides, and three polysaccharides. and three polysaccharides. b. An explanation of why carbohydrates are so soluble in water. b. An explanation of why carbohydrates are so soluble in water. c. The number of calories a gram of carbohydrates yields when oxidized. c. The number of calories a gram of carbohydrates yields when oxidized. See Lipids 1. One team of two makes a glycerol molecule; the other team of two, a molecule of Lipids palmitic acid (fatty acid). See Fig. 2.10. 1. One team of two makes a glycerol molecule; the other team of two, a molecule of 2. -

Odyssey High School Biology Vocabulary
ODYSSEY HIGH SCHOOL BIOLOGY VOCABULARY These are the vocabulary words and definitions used throughout the Biology course.They are listed in alphabetical order. Vocabulary Word Definition abiotic physical, or nonliving, factor that shapes an ecosystem abiotic factor physical, or nonliving, part of an ecosystem accurate the closeness of a measurement to the actual value acoelomate having no body cavity acoelomic lacking a body cavity acquired traits traits that develop as a result of an organism's experiences in the world the energy needed to start a reaction by exciting a stable molecule and activation energy making it a reactive molecule the energy-requiring process of moving or pumping a substance across the active transport plasma membrane against an electrical or concentration gradient (moving from low to high concentration) adapt to change to suit a new purpose a variation in a plant or animal that increases it chance of survival in its adaptation environment evolutionary divergence of members of a single phyletic line into many adaptive radiation different niches adenine purine base that pairs with thymine in DNA or uracil in RNA a small gland that produces steroid hormones, adrenaline and adrenal gland nonadrenaline, which helps control heart rate, blood pressure adventitious roots roots growing from an unusual spot, like the stem cellular process of producing energy in the form of ATP and carbon dioxide aerobic respiration from food (glucose) and oxygen; occurs in the mitochondria air the mixture of gases that make up the atmosphere -

Biomolecule Posters Working in a Group You Will Research Nucleic
Biomolecule Posters Working in a group you will research nucleic acids and create a poster to share with the class. Remember that you are teaching the class about this topic and you need to explain the topic in terms they will understand. Each group member must contribute. “Nucleic Acids” Content Describe the different functions and compositions of DNA, RNA, mRNA and tRNA. In what cellular processes are these molecules involved? Where are the nucleic acids located within the cell? Describe the composition of nucleic acids. Describe how nucleic acid polymers are constructed by dehydration synthesis and broken down into monomers by hydrolysis. How are nucleic acids related to proteins, heredity, and inheritance? Include a diagram showing the molecular structure of a nitrogenous base. An example of the molecular diagram is found to the right. Key Terms DNA (deoxyribonucleic acid), chromosome, RNA (ribonucleic acid), tRNA (transfer RNA), mRNA (mRNA), nucleotide, adenine, guanine, cytosine, thymine, uracil, double helix, sugar, phosphate, hydrogen bond, base, nucleotide, monomer, polymer Web Resources http://www.chem4kids.com/files/bio_nucleicacids.html https://www.youtube.com/watch?v=NNASRkIU5Fw http://www.davincialba.it/scienze/sc-clil-13-14/NUCLEIC_ACIDS_AND_PROTEIN_SYNTHESIS.pdf https://www.cancerquest.org/cancer-biology/gene-function http://science.howstuffworks.com/environmental/life/cellular-microscopic/nucleic-acids-info3.htm https://www.khanacademy.org/science/biology/macromolecules/nucleic-acids/a/nucleic-acids Biomolecule Posters Working in a group you will research nucleic acids and create a poster to share with the class. Remember that you are teaching the class about this topic and you need to explain the topic in terms they will understand. -

Biomolecule Presentation Working in a Group You Will Research Nucleic Acids and Create a Presentation to Share with the Class
Biomolecule Presentation Working in a group you will research nucleic acids and create a presentation to share with the class. You will present your topic to the class. Remember that you are teaching the class about this topic and you need to explain the topic in terms they will understand. Each group member must contribute. At the minimum every person in the group must design and explain at least one slide. The presentation should last between 4-10 minutes. Remember, images, graphs, tables and figures enhance your presentation. “Nucleic Acids” Content Describe the different functions and compositions of DNA, RNA, mRNA and tRNA. In what cellular processes are these molecules involved? Where are the nucleic acids located within the cell? Describe the composition of nucleic acids. Describe how nucleic acid polymers are constructed by dehydration synthesis and broken down into monomers by hydrolysis. How are nucleic acids related to proteins, heredity, and inheritance? Include a diagram showing the molecular structure of a nitrogenous base. An example of the molecular diagram is found to the right. Key Terms DNA (deoxyribonucleic acid), chromosome, RNA (ribonucleic acid), tRNA (transfer RNA), mRNA (mRNA), nucleotide, adenine, guanine, cytosine, thymine, uracil, double helix, sugar, phosphate, hydrogen bond, base, nucleotide, monomer, polymer Web Resources http://www.chem4kids.com/files/bio_nucleicacids.html https://www.youtube.com/watch?v=NNASRkIU5Fw http://www.davincialba.it/scienze/sc-clil-13-14/NUCLEIC_ACIDS_AND_PROTEIN_SYNTHESIS.pdf https://www.cancerquest.org/cancer-biology/gene-function http://science.howstuffworks.com/environmental/life/cellular-microscopic/nucleic-acids-info3.htm https://www.khanacademy.org/science/biology/macromolecules/nucleic-acids/a/nucleic-acids Biomolecule Presentation Working in a group you will research Proteins and create a presentation to share with the class. -
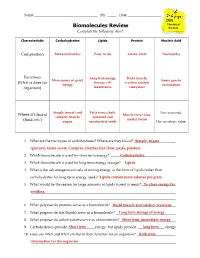
Biomolecules Review the Test
Name ___________________________________ Per. _____ Date ___________________________ DUE The day of Biomolecules Review the test. Complete the following chart: Characteristic Carbohydrates Lipids Protein Nucleic Acid Composition Monosaccharides Fatty Acids Amino Acids Nucleotides Functions Long term energy Build muscle; Main source of quick Stores genetic (What is does for storage; cell reaction catalyst energy information organism) membranes (enzymes) Simple (sweet) and Fatty tissue, both Not consumed. Where it’s found Muscle tissue (aka. complex (starch) saturated and meats); beans (food, etc.) sugars unsaturated; seeds Has no caloric value. 1. What are the two types of carbohydrates? Where are they found? _______________________Simple: sugars (________________________________glucose), tastes sweet; Complex: starc________________________________hes like flour, pasta, potatoes ________________ 2. Which biomolecule is used for short term energy? ________________________________Carbohydrates ____ 3. Which biomolecule is used for long term energy storage? ______________________________Lipids 4. What is the advantage to animals of storing energy in the form of lipids rather than carbohydrates for long-term energy needs? Lipids________________________________ contain more calories per gram__________ 5. What would be the reason for large amounts of lipids stored in seeds? ___________________To store energy for seedling________________________________ ________________________________________________ ________________________________________________________________________________