Oikos Journal
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Checklist of the Vascular Plants of Redwood National Park
Humboldt State University Digital Commons @ Humboldt State University Botanical Studies Open Educational Resources and Data 9-17-2018 Checklist of the Vascular Plants of Redwood National Park James P. Smith Jr Humboldt State University, [email protected] Follow this and additional works at: https://digitalcommons.humboldt.edu/botany_jps Part of the Botany Commons Recommended Citation Smith, James P. Jr, "Checklist of the Vascular Plants of Redwood National Park" (2018). Botanical Studies. 85. https://digitalcommons.humboldt.edu/botany_jps/85 This Flora of Northwest California-Checklists of Local Sites is brought to you for free and open access by the Open Educational Resources and Data at Digital Commons @ Humboldt State University. It has been accepted for inclusion in Botanical Studies by an authorized administrator of Digital Commons @ Humboldt State University. For more information, please contact [email protected]. A CHECKLIST OF THE VASCULAR PLANTS OF THE REDWOOD NATIONAL & STATE PARKS James P. Smith, Jr. Professor Emeritus of Botany Department of Biological Sciences Humboldt State Univerity Arcata, California 14 September 2018 The Redwood National and State Parks are located in Del Norte and Humboldt counties in coastal northwestern California. The national park was F E R N S established in 1968. In 1994, a cooperative agreement with the California Department of Parks and Recreation added Del Norte Coast, Prairie Creek, Athyriaceae – Lady Fern Family and Jedediah Smith Redwoods state parks to form a single administrative Athyrium filix-femina var. cyclosporum • northwestern lady fern unit. Together they comprise about 133,000 acres (540 km2), including 37 miles of coast line. Almost half of the remaining old growth redwood forests Blechnaceae – Deer Fern Family are protected in these four parks. -

Digitalcommons@University of Nebraska - Lincoln
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln U.S. Department of Agriculture: Agricultural Publications from USDA-ARS / UNL Faculty Research Service, Lincoln, Nebraska 1996 Bromegrasses Kenneth P. Vogel University of Nebraska-Lincoln, [email protected] K. J. Moore Iowa State University Lowell E. Moser University of Nebraska-Lincoln, [email protected] Follow this and additional works at: https://digitalcommons.unl.edu/usdaarsfacpub Vogel, Kenneth P.; Moore, K. J.; and Moser, Lowell E., "Bromegrasses" (1996). Publications from USDA- ARS / UNL Faculty. 2097. https://digitalcommons.unl.edu/usdaarsfacpub/2097 This Article is brought to you for free and open access by the U.S. Department of Agriculture: Agricultural Research Service, Lincoln, Nebraska at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Publications from USDA-ARS / UNL Faculty by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. Published 1996 17 Bromegrasses1 K.P. VOGEL USDA-ARS Lincoln, Nebraska K.J.MOORE Iowa State University Ames, Iowa LOWELL E. MOSER University of Nebraska Lincoln, Nebraska The bromegrasses belong to the genus Bromus of which there are some 100 spe cies (Gould & Shaw, 1983). The genus includes both annual and perennial cool season species adapted to temperate climates. Hitchcock (1971) described 42 bro megrass species found in the USA and Canada of which 22 were native (Gould & Shaw, 1983). Bromus is the Greek word for oat and refers to the panicle inflo rescence characteristic of the genus. The bromegrasses are C3 species (Krenzer et aI., 1975; Waller & Lewis, 1979). Of all the bromegrass species, only two are cultivated for permanent pas tures to any extent in North America. -

And Flora of the Matthaei Botanical Gardens and Nichols Arboretum
THE NATURAL COMMUNITIES AND FLORA OF THE MAttHAEI BOTANICAL GARDENS AND NICHOLS ARBORETUM BEVERLY WALTERS : MARY HEJNA : CONNIE CRANCER : JEFF PLAKKE 2011-2012 Caring for Nature, Enriching Life mbgna.umich.edu ACKNOWLEDgements This report is the product of a project entitled Assessing Globally-Ranked At-Risk Native Plant Communities: A General Conservation Survey of High Quality Natural Areas of the University of Michigan Matthaei Botanical Gardens and Nichols Arboretum, which was funded by the Institute of Museum and Library Services. Principal Investigator: Bob Grese, Director, Matthaei-Nichols. Lead Author: David Michener, Curator, Matthaei-Nichols. Editor and Project Manager: Jeff Plakke, Natural Areas Manager, Matthaei-Nichols. IMLS Sponsored Botanist: Beverly Walters, Research Museum Collection Manager (Vascular Plants), University of Michigan Herbarium. Assisting Botanist: Connie Crancer, Native Plant Specialist, Matthaei-Nichols. IMLS Sponsored GIS Technician: Mary Hejna Natural Areas Advisory Committee: Burt Barnes, Professor Emeritus, University of Michigan Dave Borneman, City of Ann Arbor Natural Areas Preservation Manager Aunita Erskine, Volunteer Steward Drew Lathin, Huron Arbor Cluster Coordinator for The Stewardship Network Kris Olson, Watershed Ecologist, Huron River Watershed Council Anton Reznicek, Assistant Director and Curator, University of Michigan Herbarium Shawn Severance, Washtenaw County Natural Areas Naturalist Sylvia Taylor, Faculty Emeritus, University of Michigan Scott Tyrell, Southeast Michigan Land Conservancy Volunteer Dana Wright, Land Stewardship Coordinator, Legacy Land Conservancy Many thanks also to Paul Berry for releasing Bev from duties at the University of Michigan Herbarium so that she could conduct the surveys, to Tony Reznicek for assistance with plant identification, and to Aunita Erskine for assistance in the field. Photographs on cover page and page 94 taken by MBGNA Staff. -

Multivariate Morphometric Study of the Bromus Erectus Group (Poaceae - Bromeae) in Slovenia
©Verlag Ferdinand Berger & Söhne Ges.m.b.H., Horn, Austria, download unter www.biologiezentrum.at Phyton (Horn, Austria) Vol. 41 Fasc. 2 295-311 28. 12. 2001 Multivariate Morphometric Study of the Bromus erectus Group (Poaceae - Bromeae) in Slovenia. By Tinka BACIC and Nejc JOGAN*) With 6 Figures Received March 15, 2001 Keywords: Gramineae, Poaceae, Bromus erectus agg. - Cluster analysis, dis- criminant analysis, principal coordinate analysis. - Flora of Slovenia. Summary BAÖIC T. & JOGAN N. 2001. Multivariate Morphometric Study of the Bromus erectus Group (Poaceae - Bromeae) in Slovenia. - Phyton (Horn, Austria) 41 (2): 295-311, 6 figures. - English with German summary. Since the end of the 19th century, six species from the Bromus erectus group have been recorded for Slovenia: B. erectus HUDSON s.str., B. transylvanicus STEUDEL, B. condensatus HACKEL, B. stenophyllus LINK, B. pannonicus KUMMER & SENDTNER and B. moellendorfianus (ASCHERSON & GRAEBNER) HAYEK. Since the delimitation of the taxa is still unclear, the purpose of the research was to investigate the dis- criminative power of distinguishing characters, used in various floristic works and also some new potentially useful distinguishing characters, subsequently to confirm whether these taxa really occur in Slovenia and finally to state their distribution. Besides the classical 'intuitive' approach to this problem, some methods of numerical taxonomy were used. The study was based on material from the herbarium LJU and second author's private collection. In the analysis 198 individuals were taken into account and 48 characters were investigated. The data were analyzed using hier- archical clustering and ordination methods (principal coordinate analysis, dis- criminant analysis) as well as some univariate statistical methods. -

(Poaceae) and Characterization
EVOLUTION AND DEVELOPMENT OF VEGETATIVE ARCHITECTURE: BROAD SCALE PATTERNS OF BRANCHING ACROSS THE GRASS FAMILY (POACEAE) AND CHARACTERIZATION OF ARCHITECTURAL DEVELOPMENT IN SETARIA VIRIDIS L. P. BEAUV. By MICHAEL P. MALAHY Bachelor of Science in Biology University of Central Oklahoma Edmond, Oklahoma 2006 Submitted to the Faculty of the Graduate College of the Oklahoma State University in partial fulfillment of the requirements for the Degree of MASTER OF SCIENCE July, 2012 EVOLUTION AND DEVELOPMENT OF VEGETATIVE ARCHITECTURE: BROAD SCALE PATTERNS OF BRANCHING ACROSS THE GRASS FAMILY (POACEAE) AND CHARACTERIZATION OF ARCHITECTURAL DEVELOPMENT IN WEEDY GREEN MILLET ( SETARIA VIRIDIS L. P. BEAUV.) Thesis Approved: Dr. Andrew Doust Thesis Adviser Dr. Mark Fishbein Dr. Linda Watson Dr. Sheryl A. Tucker Dean of the Graduate College I TABLE OF CONTENTS Chapter Page I. Evolutionary survey of vegetative branching across the grass family (poaceae) ... 1 Introduction ................................................................................................................... 1 Plant Architecture ........................................................................................................ 2 Vascular Plant Morphology ......................................................................................... 3 Grass Morphology ....................................................................................................... 4 Methods ....................................................................................................................... -

Ana Petrova, Stefan Kozuharov & Frìedrìch Ehrendorfer
Ana Petrova, Stefan Kozuharov & FrÌedrÌch Ehrendorfer Karyosystematic notes on Bromus (Gramineae), and a new species of the B. riparius polyploid complex from Bulgaria Abstract Petrova, A., Kozuharov, S. & Ehrendorfer, F.: Karyosystematic notes on Bromus (Gramineae), and a new species of the B. riparius polyploid complex from Bulgaria. - Boc conea 5: 775-780.1997. - ISSN 1120-4060. A new diploid species, Bromus parilicus, is recognized within the B. riparius aggregate and compared to B. cappadocicus (6x), B. lacmonicus (8x), and B. riparius (lOx). The decaploid B. fibrosus varo orbelicus is raised to specific rank. B. transsilvanicus probably al so belongs to the B. riparius aggregate. 2n = 14 and 2n = 70, counted on plants from N.E. Anatolia, are new chromosome numbers for B. tomentellus subsp. tomentellus and subsp. woronovii, re spectively. The first 2x count (2n = 14) for a member of the B. erectus aggregate is reported for B. moellendorffianus, which is newly recorded for Siovenia. Introduction Elaborating on our previous, preliminary work on Balkan brome-grasses (Kozuharov & al. 1981), we have eontinued our study of some plants we had the opportunity to eol leet, not only in the Balkan eountries but also in N.W. Anatolia (Mt. DIu dag). The fol- 10wing note relates new data and eonclusions eoneerning the karyology and taxonomy of several interesting perennial taxa of the Bromus riparius and, in one case (B. moellen dorffianus), the B. erectus eomplex. Live plants eolleeted in the field were nursed in the greenhouse, and seeds from wild populations were germinated in Petri dishes. Cytologieal data were obtained from the study of squash preparations of meristematie tissue from root tips dyed with Gomori's ehromie haematoxylin after fixation in Carnoy's Fluid I. -
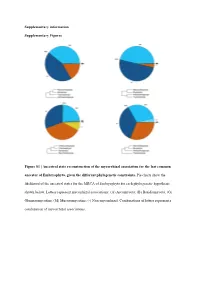
Ancestral State Reconstruction of the Mycorrhizal Association for the Last Common Ancestor of Embryophyta, Given the Different Phylogenetic Constraints
Supplementary information Supplementary Figures Figure S1 | Ancestral state reconstruction of the mycorrhizal association for the last common ancestor of Embryophyta, given the different phylogenetic constraints. Pie charts show the likelihood of the ancestral states for the MRCA of Embryophyta for each phylogenetic hypothesis shown below. Letters represent mycorrhizal associations: (A) Ascomycota; (B) Basidiomycota; (G) Glomeromycotina; (M) Mucoromycotina; (-) Non-mycorrhizal. Combinations of letters represent a combination of mycorrhizal associations. Austrocedrus chilensis Chamaecyparis obtusa Sequoiadendron giganteum Prumnopitys taxifolia Prumnopitys Prumnopitys montana Prumnopitys Prumnopitys ferruginea Prumnopitys Araucaria angustifolia Araucaria Dacrycarpus dacrydioides Dacrycarpus Taxus baccata Podocarpus oleifolius Podocarpus Afrocarpus falcatus Afrocarpus Ephedra fragilis Nymphaea alba Nymphaea Gnetum gnemon Abies alba Abies balsamea Austrobaileya scandens Austrobaileya Abies nordmanniana Thalictrum minus Thalictrum Abies homolepis Caltha palustris Caltha Abies magnifica ia repens Ranunculus Abies religiosa Ranunculus montanus Ranunculus Clematis vitalba Clematis Keteleeria davidiana Anemone patens Anemone Tsuga canadensis Vitis vinifera Vitis Tsuga mertensiana Saxifraga oppositifolia Saxifraga Larix decidua Hypericum maculatum Hypericum Larix gmelinii Phyllanthus calycinus Phyllanthus Larix kaempferi Hieronyma oblonga Hieronyma Pseudotsuga menziesii Salix reinii Salix Picea abies Salix polaris Salix Picea crassifolia Salix herbacea -

Molecular Phylogenetics of Bromus (Poaceae: Pooideae) Based on Chloroplast and Nuclear DNA Sequence Data Jeffery M
Aliso: A Journal of Systematic and Evolutionary Botany Volume 23 | Issue 1 Article 35 2007 Molecular Phylogenetics of Bromus (Poaceae: Pooideae) Based on Chloroplast and Nuclear DNA Sequence Data Jeffery M. Saarela University of British Columbia, Vancouver, British Columbia, Canada Paul M. Peterson National Museum of Natural History, Smithsonian Institution, Washington, D.C. Ryan M. Keane Imperial College of Science, Technology and Medicine, Ascot, UK Jacques Cayouette Eastern Cereal and Oilseed Research Centre, Agriculture and Agri-Food Canada, Ottawa, Ontario, Canada Sean W. Graham University of British Columbia, Vancouver, British Columbia, Canada Follow this and additional works at: http://scholarship.claremont.edu/aliso Part of the Botany Commons, and the Ecology and Evolutionary Biology Commons Recommended Citation Saarela, Jeffery M.; Peterson, Paul M.; Keane, Ryan M.; Cayouette, Jacques; and Graham, Sean W. (2007) "Molecular Phylogenetics of Bromus (Poaceae: Pooideae) Based on Chloroplast and Nuclear DNA Sequence Data," Aliso: A Journal of Systematic and Evolutionary Botany: Vol. 23: Iss. 1, Article 35. Available at: http://scholarship.claremont.edu/aliso/vol23/iss1/35 Aliso 23, pp. 450–467 ᭧ 2007, Rancho Santa Ana Botanic Garden MOLECULAR PHYLOGENETICS OF BROMUS (POACEAE: POOIDEAE) BASED ON CHLOROPLAST AND NUCLEAR DNA SEQUENCE DATA JEFFERY M. SAARELA,1,5 PAUL M. PETERSON,2 RYAN M. KEANE,3 JACQUES CAYOUETTE,4 AND SEAN W. GRAHAM1 1Department of Botany and UBC Botanical Garden and Centre for Plant Research, University of British Columbia, Vancouver, British Columbia, V6T 1Z4, Canada; 2Department of Botany, National Museum of Natural History, MRC- 166, Smithsonian Institution, Washington, D.C. 20013-7012, USA; 3Department of Biology, Imperial College of Science, Technology and Medicine, Silwood Park, Ascot, Berkshire, SL5 7PY, UK; 4Eastern Cereal and Oilseed Research Centre (ECORC), Agriculture and Agri-Food Canada, Central Experimental Farm, Wm. -

Checklist of Vascular Plants of the Southern Rocky Mountain Region
Checklist of Vascular Plants of the Southern Rocky Mountain Region (VERSION 3) NEIL SNOW Herbarium Pacificum Bernice P. Bishop Museum 1525 Bernice Street Honolulu, HI 96817 [email protected] Suggested citation: Snow, N. 2009. Checklist of Vascular Plants of the Southern Rocky Mountain Region (Version 3). 316 pp. Retrievable from the Colorado Native Plant Society (http://www.conps.org/plant_lists.html). The author retains the rights irrespective of its electronic posting. Please circulate freely. 1 Snow, N. January 2009. Checklist of Vascular Plants of the Southern Rocky Mountain Region. (Version 3). Dedication To all who work on behalf of the conservation of species and ecosystems. Abbreviated Table of Contents Fern Allies and Ferns.........................................................................................................12 Gymnopserms ....................................................................................................................19 Angiosperms ......................................................................................................................21 Amaranthaceae ............................................................................................................23 Apiaceae ......................................................................................................................31 Asteraceae....................................................................................................................38 Boraginaceae ...............................................................................................................98 -

Southern Ontario Vascular Plant Species List
Southern Ontario Vascular Plant Species List (Sorted by Scientific Name) Based on the Ontario Plant List (Newmaster et al. 1998) David J. Bradley Southern Science & Information Section Ontario Ministry of Natural Resources Peterborough, Ontario Revised Edition, 2007 Southern Ontario Vascular Plant Species List This species checklist has been compiled in order to assist field biologists who are sampling vegetative plots in Southern Ontario. It is not intended to be a complete species list for the region. The intended range for this vascular plant list is Ecoregions (Site Regions) 5E, 6E and 7E. i Nomenclature The nomenclature used for this listing of 2,532 plant species, subspecies and varieties, is in accordance with the Ontario Plant List (OPL), 1998 [see Further Reading for full citation]. This is the Ontario Ministry of Natural Resource’s publication which has been selected as the corporate standard for plant nomenclature. There have been many nomenclatural innovations in the past several years since the publication of the Ontario Plant List that are not reflected in this listing. However, the OPL has a listing of many of the synonyms that have been used recently in the botanical literature. For a more up to date listing of scientific plant names visit either of the following web sites: Flora of North America - http://www.efloras.org/flora_page.aspx?flora_id=1 NatureServe - http://www.natureserve.org/explorer/servlet/NatureServe?init=Species People who are familiar with the Natural Heritage Information Centre (NHIC) plant species list for Ontario, will notice some changes in the nomenclature. For example, most of the Aster species have now been put into the genus Symphyotrichum, with a few into the genus Eurybia. -

Poaceae), and the Interrelation of Whole-Genome Duplication and Evolutionary Radiations in This Grass Tribe
bioRxiv preprint doi: https://doi.org/10.1101/2020.06.05.129320; this version posted June 6, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Molecular phylogenetics and micromorphology of Australasian Stipeae (Poaceae), and the interrelation of whole-genome duplication and evolutionary radiations in this grass tribe Natalia Tkach,1 Marcin Nobis,2 Julia Schneider,1 Hannes Becher,1,3 Grit Winterfeld,1 Mary E. Barkworth,4 Surrey W. L. Jacobs5, Martin Röser1 1 Martin Luther University Halle-Wittenberg, Institute of Biology, Geobotany and Botanical Garden, Dept. of Systematic Botany, Neuwerk 21, 06108 Halle, Germany 2 Institute of Botany, Faculty of Biology, Jagiellonian University, Gronostajowa 3 st., 30-387 Kraków, Poland 3 present address: Institute of Evolutionary Biology, School of Biological Sciences, University of Edinburgh, Charlotte Auerbach Road, Edinburgh EH9 3FL, UK 4 Biology Department, Utah State University, 5305 Old Main Hill, Logan Utah, 84322, USA 5 deceased Addresses for correspondence: Martin Röser, [email protected]; Natalia Tkach, [email protected] ABSTRACT The mainly Australian grass genus Austrostipa with ca. 64 species represents a remarkable example of an evolutionary radiation. To investigate aspects of diversification, macro- and micromorphological variation in this genus we conducted a molecular phylogenetic and scanning electron microscopy (SEM) analysis including representatives from all of its accepted subgenera. Plastid DNA variation within Austrostipa was low and only few lineages were resolved. -

A New Species of Perennial Bromus (Bromeae, Poaceae) from the Iberian Peninsula
A peer-reviewed open-access journal PhytoKeys 121: 1–12 (2019)A new species of perennial Bromus from the Iberian Peninsula 1 doi: 10.3897/phytokeys.121.32588 RESEARCH ARTICLE http://phytokeys.pensoft.net Launched to accelerate biodiversity research A new species of perennial Bromus (Bromeae, Poaceae) from the Iberian Peninsula Carmen Acedo1, Félix Llamas1 1 Research Group Taxonomy and Conservation of Biodiversity TaCoBi, Department of Biodiversity and Envi- ronment Management, University of León, E-24071. León, Spain Corresponding author: Carmen Acedo ([email protected]) Academic editor: M. Vorontsova | Received 20 December 2018 | Accepted 22 March 2019 | Published 24 April 2019 Citation: Acedo C, Llamas F (2019) A new species of perennial Bromus (Bromeae, Poaceae) from the Iberian Peninsula. PhytoKeys 121: 1–12. https://doi.org/10.3897/phytokeys.121.32588 Abstract During a survey of the genus Bromus for the ongoing Flora Iberica, B. picoeuropeanus sp. nov., a new orophilous species of perennial Bromus from Picos de Europa National Park, was found, and it is described and illustrated here. This new species belongs to the Bromus erectus complex and differs from the other perennial species of this group occurring in the Iberian Peninsula in its well-developed rhizome, the small innovation leaves and all peduncles and branches shorter than the spikelets. B. picoeuropeanus grows on calcareous stony soils associated with dry places. We provide a description and illustrations of the new spe- cies and an identification key for the most related European perennial species belonging to the complex. Keywords Bromea, Bromus subg. Festucoides, Bromus erectus complex, Bromus picoeuropeanus, Cantabrian range, Identification Key, New species, Poaceae, Spain, Taxonomy Introduction The genus Bromus L.