And HPV18-Infected Early Stage Cervical Cancers and Normal
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Propranolol-Mediated Attenuation of MMP-9 Excretion in Infants with Hemangiomas
Supplementary Online Content Thaivalappil S, Bauman N, Saieg A, Movius E, Brown KJ, Preciado D. Propranolol-mediated attenuation of MMP-9 excretion in infants with hemangiomas. JAMA Otolaryngol Head Neck Surg. doi:10.1001/jamaoto.2013.4773 eTable. List of All of the Proteins Identified by Proteomics This supplementary material has been provided by the authors to give readers additional information about their work. © 2013 American Medical Association. All rights reserved. Downloaded From: https://jamanetwork.com/ on 10/01/2021 eTable. List of All of the Proteins Identified by Proteomics Protein Name Prop 12 mo/4 Pred 12 mo/4 Δ Prop to Pred mo mo Myeloperoxidase OS=Homo sapiens GN=MPO 26.00 143.00 ‐117.00 Lactotransferrin OS=Homo sapiens GN=LTF 114.00 205.50 ‐91.50 Matrix metalloproteinase‐9 OS=Homo sapiens GN=MMP9 5.00 36.00 ‐31.00 Neutrophil elastase OS=Homo sapiens GN=ELANE 24.00 48.00 ‐24.00 Bleomycin hydrolase OS=Homo sapiens GN=BLMH 3.00 25.00 ‐22.00 CAP7_HUMAN Azurocidin OS=Homo sapiens GN=AZU1 PE=1 SV=3 4.00 26.00 ‐22.00 S10A8_HUMAN Protein S100‐A8 OS=Homo sapiens GN=S100A8 PE=1 14.67 30.50 ‐15.83 SV=1 IL1F9_HUMAN Interleukin‐1 family member 9 OS=Homo sapiens 1.00 15.00 ‐14.00 GN=IL1F9 PE=1 SV=1 MUC5B_HUMAN Mucin‐5B OS=Homo sapiens GN=MUC5B PE=1 SV=3 2.00 14.00 ‐12.00 MUC4_HUMAN Mucin‐4 OS=Homo sapiens GN=MUC4 PE=1 SV=3 1.00 12.00 ‐11.00 HRG_HUMAN Histidine‐rich glycoprotein OS=Homo sapiens GN=HRG 1.00 12.00 ‐11.00 PE=1 SV=1 TKT_HUMAN Transketolase OS=Homo sapiens GN=TKT PE=1 SV=3 17.00 28.00 ‐11.00 CATG_HUMAN Cathepsin G OS=Homo -

Enzymatic Encoding Methods for Efficient Synthesis Of
(19) TZZ__T (11) EP 1 957 644 B1 (12) EUROPEAN PATENT SPECIFICATION (45) Date of publication and mention (51) Int Cl.: of the grant of the patent: C12N 15/10 (2006.01) C12Q 1/68 (2006.01) 01.12.2010 Bulletin 2010/48 C40B 40/06 (2006.01) C40B 50/06 (2006.01) (21) Application number: 06818144.5 (86) International application number: PCT/DK2006/000685 (22) Date of filing: 01.12.2006 (87) International publication number: WO 2007/062664 (07.06.2007 Gazette 2007/23) (54) ENZYMATIC ENCODING METHODS FOR EFFICIENT SYNTHESIS OF LARGE LIBRARIES ENZYMVERMITTELNDE KODIERUNGSMETHODEN FÜR EINE EFFIZIENTE SYNTHESE VON GROSSEN BIBLIOTHEKEN PROCEDES DE CODAGE ENZYMATIQUE DESTINES A LA SYNTHESE EFFICACE DE BIBLIOTHEQUES IMPORTANTES (84) Designated Contracting States: • GOLDBECH, Anne AT BE BG CH CY CZ DE DK EE ES FI FR GB GR DK-2200 Copenhagen N (DK) HU IE IS IT LI LT LU LV MC NL PL PT RO SE SI • DE LEON, Daen SK TR DK-2300 Copenhagen S (DK) Designated Extension States: • KALDOR, Ditte Kievsmose AL BA HR MK RS DK-2880 Bagsvaerd (DK) • SLØK, Frank Abilgaard (30) Priority: 01.12.2005 DK 200501704 DK-3450 Allerød (DK) 02.12.2005 US 741490 P • HUSEMOEN, Birgitte Nystrup DK-2500 Valby (DK) (43) Date of publication of application: • DOLBERG, Johannes 20.08.2008 Bulletin 2008/34 DK-1674 Copenhagen V (DK) • JENSEN, Kim Birkebæk (73) Proprietor: Nuevolution A/S DK-2610 Rødovre (DK) 2100 Copenhagen 0 (DK) • PETERSEN, Lene DK-2100 Copenhagen Ø (DK) (72) Inventors: • NØRREGAARD-MADSEN, Mads • FRANCH, Thomas DK-3460 Birkerød (DK) DK-3070 Snekkersten (DK) • GODSKESEN, -

Tnfa-Induced Mucin 4 Expression Elicits Trastuzumab Resistance in HER2-Positive Breast Cancer María F
Published OnlineFirst October 3, 2016; DOI: 10.1158/1078-0432.CCR-16-0970 Cancer Therapy: Clinical Clinical Cancer Research TNFa-Induced Mucin 4 Expression Elicits Trastuzumab Resistance in HER2-Positive Breast Cancer María F. Mercogliano1, Mara De Martino1, Leandro Venturutti1, Martín A. Rivas2, Cecilia J. Proietti1, Gloria Inurrigarro3, Isabel Frahm3, Daniel H. Allemand4, Ernesto Gil Deza5, Sandra Ares5, Felipe G. Gercovich5, Pablo Guzman 6, Juan C. Roa6,7, Patricia V. Elizalde1, and Roxana Schillaci1 Abstract Purpose: Although trastuzumab administration improved the Results: TNFa overexpression turned trastuzumab-sensitive outcome of HER2-positive breast cancer patients, resistance cells and tumors into resistant ones. Histopathologic findings events hamper its clinical benefits. We demonstrated that TNFa revealed mucin foci in TNFa-producing tumors. TNFa induced stimulation in vitro induces trastuzumab resistance in HER2- upregulation of MUC4 that reduced trastuzumab binding to its positive breast cancer cell lines. Here, we explored the mechanism epitope and impaired ADCC. Silencing MUC4 enhanced trastu- of TNFa-induced trastuzumab resistance and the therapeutic zumab binding, increased ADCC, and overcame trastuzumab and strategies to overcome it. trastuzumab-emtansine antiproliferative effects in TNFa-overex- Experimental Design: Trastuzumab-sensitive breast cancer pressing cells. Accordingly, administration of TNFa-blocking cells, genetically engineered to stably overexpress TNFa,and antibodies downregulated MUC4 and sensitized de novo trastu- de novo trastuzumab-resistant tumors, were used to evaluate zumab-resistant breast cancer cells and tumors to trastuzumab. In trastuzumab response and TNFa-blocking antibodies effective- HER2-positive breast cancer samples, MUC4 expression was ness respectively. Immunohistochemistry and antibody-depen- found to be an independent predictor of poor disease-free survival dent cell cytotoxicity (ADCC), together with siRNA strategy, (P ¼ 0.008). -

A Stealth Cloak for Cancer Cells
BMB Rep. 2021; 54(7): 344-355 BMB www.bmbreports.org Reports Invited Mini Review Mucin in cancer: a stealth cloak for cancer cells Dong-Han Wi1, Jong-Ho Cha2,3 & Youn-Sang Jung1,* 1Department of Life Science, Chung-Ang University, Seoul, 06974, 2Department of Biomedical Sciences, College of Medicine, Inha University, Incheon 22212, 3Department of Biomedical Science, Program in Biomedical Science and Engineering, Graduate school, Inha University, Incheon 22212, Korea Mucins are high molecular-weight epithelial glycoproteins and mucinous colorectal carcinoma (MCC) (3). Since tumor growth are implicated in many physiological processes, including epit- sites induce inhospitable conditions for them to survive, helial cell protection, signaling transduction, and tissue home- mucins are suggested as an oncogenic microenvironment that ostasis. Abnormality of mucus expression and structure contri- avoids hypoxia, acidic, and other biological hurdles. The com- butes to biological properties related to human cancer progress- position and structure of mucins enable them to mimic the ion. Tumor growth sites induce inhospitable conditions. Many surface of tumor cells like the surface of normal epithelial cells kinds of research suggest that mucins provide a microenviron- (4). Additionally, the mucus layer captures growth factors or ment to avoid hypoxia, acidic, and other biological conditions cytokines, contributing to cell growth of the tumor. Alter- that promote cancer progression. Given that the mucus layer natively, these properties interfere with the interaction bet- captures growth factors or cytokines, we propose that mucin ween the immune system and tumor cells. Indeed, a high helps to ameliorate inhospitable conditions in tumor-growing concentration of soluble mucins downregulates the motility sites. -

Gephyrin: a Scaffold That Builds a Phase at the Inhibitory Postsynapses
www.nature.com/cr www.cell-research.com RESEARCH HIGHLIGHT OPEN Gephyrin: a scaffold that builds a phase at the inhibitory postsynapses Christian Hoffmann 1 and Dragomir Milovanovic 1 Cell Research (2021) 31:245–246; https://doi.org/10.1038/s41422-020-00440-2 The scaffolding protein gephyrin is a core component of many GPHN-E. Besides GlyR-β, TM3–4 loops of GABAAR α3-subunit also inhibitory synapses. Bai and colleagues now show that phase phase separates with GPHN-E suggesting liquid condensation to separation of gephyrin with the subunits of both glycine and be a general feature of inhibitory synapses (Fig. 1). The phase GABA receptors underlies the formation of postsynaptic separation occurs at the equimolar ratio of receptors and scaffold sheets at the inhibitory synapses. proteins, a range similar to their physiological concentrations.8 To assure precise processing of information, contacts between There are several implications of this study. First, the observa- neurons require the alignment of the release sites, presynaptic tion that different regions of the same proteins (GlyR-β and GPHN- terminals, with the ionotropic transmitter receptors at the E) are important for phase separation or direct binding, suggests postsynaptic plasma membrane. Downstream of these receptors, that these two processes could be regulated separately. The scaffold molecules, kinases and cytoskeleton components all positively charged region of GlyR-β (a.a. 354–383) and negatively orchestrate the signaling cascade induced by the neurotransmitter charged surface at the subdomain II of GPHN-E are essential for fl 1234567890();,: binding and ion in ux. Each neuron in the mammalian central condensate formation, despite hydrophobic interactions being nervous system relies on both the excitatory and inhibitory inputs. -

Digitalcommons@UNMC Regulation of the Transmembrane Mucin MUC4
University of Nebraska Medical Center DigitalCommons@UNMC Theses & Dissertations Graduate Studies Fall 12-18-2015 Regulation of the transmembrane mucin MUC4 by Wnt/β-catenin in gastrointestinal cancers Priya Pai University of Nebraska Medical Center Follow this and additional works at: https://digitalcommons.unmc.edu/etd Part of the Biochemistry Commons, and the Molecular Biology Commons Recommended Citation Pai, Priya, "Regulation of the transmembrane mucin MUC4 by Wnt/β-catenin in gastrointestinal cancers" (2015). Theses & Dissertations. 58. https://digitalcommons.unmc.edu/etd/58 This Dissertation is brought to you for free and open access by the Graduate Studies at DigitalCommons@UNMC. It has been accepted for inclusion in Theses & Dissertations by an authorized administrator of DigitalCommons@UNMC. For more information, please contact [email protected]. i Regulation of the transmembrane mucin MUC4 by Wnt/β- catenin in gastrointestinal cancers By PRIYA PAI A DISSERTATION Presented to the Faculty of The University of Nebraska Graduate College In Partial Fulfillment of the Requirements For the Degree of Doctor of Philosophy Department of Biochemistry and Molecular Biology Graduate Program Under the Supervision of Professor Surinder K. Batra University of Nebraska Medical Center Omaha, Nebraska November, 2015 ii Regulation of the transmembrane mucin MUC4 by Wnt/β-catenin in gastrointestinal cancers Priya Pai, PhD. University of Nebraska Medical Center, 2015 Supervisor: Surinder K. Batra, PhD. The transmembrane mucin MUC4 is a high molecular weight glycoprotein that is expressed de novo in pancreatic ductal adenocarcinoma (PDAC). MUC4 has been shown to play a tumor-promoting role in malignancies such as PDAC, ovarian cancer and breast cancer. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -
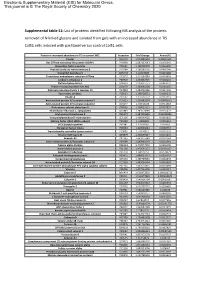
Supplemental Table S1
Electronic Supplementary Material (ESI) for Molecular Omics. This journal is © The Royal Society of Chemistry 2020 Supplemental table S1: List of proteins identified following MS analysis of the proteins removed of N-linked glycans and isolated from gels with an increased abundance in TIS Cal51 cells induced with paclitaxel versus control Cal51 cells. Protein in increased abundance in TIS vs control WCL Accession Fold Change Anova (P) Plectin Q15149 1.073855593 0.00691631 Ras GTPase-activating-like protein IQGAP1 P46940 1.087337643 0.0176342 Elongation factor1-gamma P26641 1.138709703 0.0116496 Peptidyl-prolyl cis-transisomerase B P23284 1.188383105 0.0436246 Dipeptidyl peptidase 3 Q9NY33 1.20163605 0.0215448 Transitional endoplasmic reticulum ATPase P55072 1.214194884 0.0449691 Carbonic anhydrase 2 P00918 1.232852325 0.0158141 Clathrin heavy chain 1 Q00610 1.239621773 0.0463237 Protein transport protein Sec 31A O94979 1.263565104 0.0284155 Aldo-ketoreductase family 1 member C1 Q04828 1.282092186 0.0324406 Spermidine synthase P19623 1.298728621 0.0196232 Plastin-3 P13797 1.310756772 0.0161319 Actin-related protein 2/3 complex subunit 5 O15511 1.333483524 0.00476923 Actin-related protein 2/3 complex subunit 2 O15144 1.35416168 0.0411018 Proteasome subunit alpha type-5 P28066 1.358015551 0.0337657 Thioredoxin reductase 1, cytoplasmic Q16881 1.383670089 0.0235472 Acyl-protein thioesterase 2 O95372 1.387415589 0.00233899 Isoaspartylpeptidase/L-asparaginase Q7L266 1.408149002 0.0319602 Splicing factor U2AF 65kDa subunit P26368 1.41489991 0.0256619 -

Essential Trace Elements in Human Health: a Physician's View
Margarita G. Skalnaya, Anatoly V. Skalny ESSENTIAL TRACE ELEMENTS IN HUMAN HEALTH: A PHYSICIAN'S VIEW Reviewers: Philippe Collery, M.D., Ph.D. Ivan V. Radysh, M.D., Ph.D., D.Sc. Tomsk Publishing House of Tomsk State University 2018 2 Essential trace elements in human health UDK 612:577.1 LBC 52.57 S66 Skalnaya Margarita G., Skalny Anatoly V. S66 Essential trace elements in human health: a physician's view. – Tomsk : Publishing House of Tomsk State University, 2018. – 224 p. ISBN 978-5-94621-683-8 Disturbances in trace element homeostasis may result in the development of pathologic states and diseases. The most characteristic patterns of a modern human being are deficiency of essential and excess of toxic trace elements. Such a deficiency frequently occurs due to insufficient trace element content in diets or increased requirements of an organism. All these changes of trace element homeostasis form an individual trace element portrait of a person. Consequently, impaired balance of every trace element should be analyzed in the view of other patterns of trace element portrait. Only personalized approach to diagnosis can meet these requirements and result in successful treatment. Effective management and timely diagnosis of trace element deficiency and toxicity may occur only in the case of adequate assessment of trace element status of every individual based on recent data on trace element metabolism. Therefore, the most recent basic data on participation of essential trace elements in physiological processes, metabolism, routes and volumes of entering to the body, relation to various diseases, medical applications with a special focus on iron (Fe), copper (Cu), manganese (Mn), zinc (Zn), selenium (Se), iodine (I), cobalt (Co), chromium, and molybdenum (Mo) are reviewed. -

Serial Analysis of Gene Expression in Normal P53 Null Mammary Epithelium
Oncogene (2002) 21, 6366 – 6376 ª 2002 Nature Publishing Group All rights reserved 0950 – 9232/02 $25.00 www.nature.com/onc Serial analysis of gene expression in normal p53 null mammary epithelium C Marcelo Aldaz*,1, Yuhui Hu1, Rachael Daniel1, Sally Gaddis1, Frances Kittrell2 and Daniel Medina2 1The University of Texas M.D. Anderson Cancer Center, Department of Carcinogenesis, Smithville, Texas, TX 78957, USA; 2Baylor College of Medicine Department of Molecular and Cellular Biology, Houston, Texas, TX 77030, USA Much evidence has accumulated implicating the p53 gene function although activating mutations were also as of importance in breast carcinogenesis. However, observed. Usually p53 abnormalities associate with much still remains to be uncovered on the specific poorer clinical outcome. This, likely, is the consequence downstream pathways influenced by this important of the known critical roles of p53 in regulating the cell activator/repressor of transcription. This study investi- cycle, apoptosis, DNA repair and maintaining genome gated the effects of a p53 null genotype on the stability (Levine, 1997). The loss of wild type p53 transcriptome of ‘normal’ mouse mammary epithelium function is clearly an important event in breast using a unique in vivo model of preneoplastic transforma- tumorigenesis as documented both in human and murine tion. We used SAGE for the comparative analysis of p53 systems (Donehower et al., 1995; Elledge and Allred, wild type (wt) and null mammary epithelium unexposed 1994). However, the exact mechanisms by which such and exposed to hormonal stimulation. Analysis of the lack of normal gene function leads to cancer formation hormone exposed samples provided a comprehensive view and progression are only beginning to be understood. -

In the MOCS2 Gene
Novel variant (c.472_477del) in the MOCS2 gene Aleksandra Jezela-Stanek1, Witold Blaz2, Artur Gora3, Malgorzata Bochenska2, Katarzyna Kusmierska4, and Jolanta Sykut-Cegielska4 1National Tuberculosis and Lung Diseases Institute 2Saint Jadwiga the Queen Clinical Provincial Hospital No2 3Tunneling Group, Biotechnology Centre, Silesian University of Technology 4Institute of Mother and Child May 26, 2020 Abstract Molybdenum cofactor deficiency type B (MOCODB, #252160) is a rare autosomal recessive metabolic disorder characterized by intractable seizures of neonatal-onset, muscular spasticity, accompanying with hypouricemia, elevated urinary sulfite levels and craniofacial dysmorphism. Thirty-five patients were reported to date. Our paper aimed to delineate the disease genotype by presenting another patient, in whom novel, inframe variant within the MOCS2 gene was identified. Its clinical significance was supported by the medical history and analysis of the possible mutation consequences on a molecular level with the use of the available crystal structure of the human molybdopterin synthase complex. Moreover, potential pathomechanism resulting from molecular defect was presented, giving original insight into current knowledge on this rare disease, including treatment options. Introduction Molybdenum cofactor deficiency type B (MOCODB, #252160) is an autosomal recessive metabolic dis- order characterized by intractable seizures of neonatal-onset, muscular spasticity, accompanying with hy- pouricemia, elevated urinary sulfite levels and craniofacial dysmorphism. It came to medical attention first in 1980 (Johnson, 1980). Affected children show severe neurologic complications, which may lead to early death, rarely (only three cases described to date) presented with a milder form with global developmen- tal delay without seizures (Arican, 2019). The disorder results from decreased activity of sulfite oxidase (SUOX; EC 1.8.3.1) and xanthine dehydrogenase (XDH; EC 1.17.1.4 and 1.17.3.2), which are molybdenum cofactor-dependent for their activity. -

Proteomic Analysis of Cancer and Mesothelial Cells Reveals an Increase in Mucin 5AC During Ovarian Cancer and Peritoneal Interaction
JOURNAL OF PROTEOMICS 103 (2014) 204– 215 Available online at www.sciencedirect.com ScienceDirect www.elsevier.com/locate/jprot Proteomic analysis of cancer and mesothelial cells reveals an increase in Mucin 5AC during ovarian cancer and peritoneal interaction Natasha Musrapa,b, George S. Karagiannisa,b, Punit Saraona,b, Ihor Batruchb, Chris Smithc, Eleftherios P. Diamandisa,b,c,⁎ aDepartment of Laboratory Medicine and Pathobiology, University of Toronto, Toronto, Ontario, Canada bDepartment of Pathology and Laboratory Medicine, Mount Sinai Hospital, Toronto, Ontario, Canada cDepartment of Clinical Biochemistry, University Health Network, Toronto, Ontario, Canada ARTICLE INFO ABSTRACT Article history: Ovarian cancer is a highly metastatic disease that is often characterized by widespread Received 4 December 2013 abdominal dissemination. A hallmark of ovarian cancer progression is the attachment of Accepted 27 March 2014 malignant cells to the mesothelium and the formation of invasive peritoneal implants. Available online 12 April 2014 Therefore, delineating factors involved in cancer-peritoneal cell interaction is critical to improving patient survival, as it may lead to the discovery of novel therapeutic targets. As Keywords: such, we aimed to identify proteins that participate in this interaction by comparing the Ovarian cancer secreted proteome of a co-culture model containing ovarian cancer (OVCAR-5) and mesothelial Proteomics cells (LP-9), to their respective monoculture secretomes. In total, 49 proteins were differentially Co-culture secreted during cancer and mesothelial cell contact. Relative mRNA expression of candidates Peritoneum was performed, which revealed a significant increase in MUC5AC gene expression in cancer Metastasis cells cultured in three different co-culture models (OVCAR-5 and LP-9; BG-1 and LP-9; OV-90 and Mucin 5AC LP-9).