Title Purification and Properties of L-Methionine Decarboxylase Of
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Surfen, a Small Molecule Antagonist of Heparan Sulfate
Surfen, a small molecule antagonist of heparan sulfate Manuela Schuksz*†, Mark M. Fuster‡, Jillian R. Brown§, Brett E. Crawford§, David P. Ditto¶, Roger Lawrence*, Charles A. Glass§, Lianchun Wang*, Yitzhak Torʈ, and Jeffrey D. Esko*,** *Department of Cellular and Molecular Medicine, Glycobiology Research and Training Center, †Biomedical Sciences Graduate Program, ‡Department of Medicine, Division of Pulmonary and Critical Care Medicine and Veteran’s Administration San Diego Medical Center, ¶Moores Cancer Center, and ʈDepartment of Chemistry and Biochemistry, University of California at San Diego, La Jolla, CA 92093; and §Zacharon Pharmaceuticals, Inc, 505 Coast Blvd, South, La Jolla, CA 92037 Communicated by Carolyn R. Bertozzi, University of California, Berkeley, CA, June 18, 2008 (received for review May 26, 2007) In a search for small molecule antagonists of heparan sulfate, Surfen (bis-2-methyl-4-amino-quinolyl-6-carbamide) was first we examined the activity of bis-2-methyl-4-amino-quinolyl-6- described in 1938 as an excipient for the production of depot carbamide, also known as surfen. Fluorescence-based titrations insulin (16). Subsequent studies have shown that surfen can indicated that surfen bound to glycosaminoglycans, and the extent block C5a receptor binding (17) and lethal factor (LF) produced of binding increased according to charge density in the order by anthrax (18). It was also reported to have modest heparin- heparin > dermatan sulfate > heparan sulfate > chondroitin neutralizing effects in an oral feeding experiments in rats (19), sulfate. All charged groups in heparin (N-sulfates, O-sulfates, and but to our knowledge, no further studies involving heparin have carboxyl groups) contributed to binding, consistent with the idea been conducted, and its effects on HS are completely unknown. -

Polyamine Biosynthesis in Human Fetal Liver and Brain
Pediat. Res. 8: 231-237 (1974) S-Adenosylmethionine decarboxylase polyamines developmental biochemistry putrescine fetus spermidine ornithine decarboxylase Polyamine Biosynthesis in Human Fetal Liver and Brain JOHN A. STURMAN'~~]AND GERALD E. GAULL Department of Pediatric Research, Institute for Basic liesearch in Mental Retardation, Staten Island, and Department of Pediatrics, Division of Medical Genetics and Clinical Genetics Center, Mount Sinai School of Medicine of the City University of New York, New York, New York, USA Extract S-Adenosylmethionine decarboxylase specific activity in fetal (9.0-25.0 cm crown- rump length) human liver was 20-fold greater than in mature human liver, both in the absence of putrescine (63.1 f 7.2 versus 3.5 =t 0.8 pmol C02/mgprotein/30 min) or in the presence of putrescine (1 16.9 + 8.2 uersus 6.2 =I= 1.0 pmol C02/mg protein/30 min). Extracts of fetal human brain did not have a significantly greater specific ac- tivity of S-adenosylmethionine decarboxylase than extracts of mature human brain when assayed in the absence of putrescine and had less when assayed in the presence of putrescine (84.7 =t 14.9 versus 175.5 f 30.6 pmol C02/mgprotein/30 min). Ornithine decarboxylase specific activity was greater in fetal liver than in mature liver (8.8 =t1.6 versus 1.1 f 0.3 pmol CO 2/mg protein/30 min) and 20-fold greater in fetal brain than in mature brain (7 1.2 =t 12.1 uersus 3.1 =I= 1.4 pmol CO z/mg protein/30 min). -
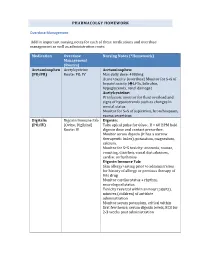
PHARMACOLGY HOMEWORK Overdose Management Add In
PHARMACOLGY HOMEWORK Overdose Management Add in important nursing notes for each of these medications and overdose management as well as administration route. Medication Overdose Nursing Notes (*Homework) Management (Routes) Acetaminophen Acetylcysteine Acetaminophen: (PO/PR) Route: PO, IV Max daily dose: 4000mg Acute toxicity (overdose) Monitor for S+S of hepatotoxicity (éLFTs, bilirubin, hypoglycemia, renal damage) Acetylcysteine: IV infusion: monitor for fluid overload and signs of hyponatremia such as changes in mental status Monitor for S+S of aspiration, bronchospasm, excess secretions Digitalis Digoxin Immune Fab Digoxin: (PO/IV) (Ovine, Digibind) Take apical pulse for 60sec. If < 60 BPM hold Route: IV digoxin dose and contact prescriber. Monitor serum digoxin (it has a narrow therapeutic index), potassium, magnesium, calcium. Monitor for S+S toxicity: anorexia, nausea, vomiting, diarrhea, visual disturbances, cardiac arrhythmias Digoxin Immune Fab: Skin allergy testing prior to administration for history of allergy or previous therapy of this drug Monitor cardiac status + rhythm, neurological status Toxicity reversal within an hour (adults), minutes (children) of antidote administration Monitor serum potassium, critical within first few hours; serum digoxin levels, ECG for 2-3 weeks post administration Heparin Protamine sulfate Heparin: (IV/SC) (IV) Monitor for spontaneous bleeding, thrombocytopenia Monitor aPTT levels Protamine Sulfate: Sudden drop in BP Monitor BP+ P q15-30 min Monitor aPTT Opioid Naloxone (IV-adults, Opioids: -

Jp Xvii the Japanese Pharmacopoeia
JP XVII THE JAPANESE PHARMACOPOEIA SEVENTEENTH EDITION Official from April 1, 2016 English Version THE MINISTRY OF HEALTH, LABOUR AND WELFARE Notice: This English Version of the Japanese Pharmacopoeia is published for the convenience of users unfamiliar with the Japanese language. When and if any discrepancy arises between the Japanese original and its English translation, the former is authentic. The Ministry of Health, Labour and Welfare Ministerial Notification No. 64 Pursuant to Paragraph 1, Article 41 of the Law on Securing Quality, Efficacy and Safety of Products including Pharmaceuticals and Medical Devices (Law No. 145, 1960), the Japanese Pharmacopoeia (Ministerial Notification No. 65, 2011), which has been established as follows*, shall be applied on April 1, 2016. However, in the case of drugs which are listed in the Pharmacopoeia (hereinafter referred to as ``previ- ous Pharmacopoeia'') [limited to those listed in the Japanese Pharmacopoeia whose standards are changed in accordance with this notification (hereinafter referred to as ``new Pharmacopoeia'')] and have been approved as of April 1, 2016 as prescribed under Paragraph 1, Article 14 of the same law [including drugs the Minister of Health, Labour and Welfare specifies (the Ministry of Health and Welfare Ministerial Notification No. 104, 1994) as of March 31, 2016 as those exempted from marketing approval pursuant to Paragraph 1, Article 14 of the Same Law (hereinafter referred to as ``drugs exempted from approval'')], the Name and Standards established in the previous Pharmacopoeia (limited to part of the Name and Standards for the drugs concerned) may be accepted to conform to the Name and Standards established in the new Pharmacopoeia before and on September 30, 2017. -

Estonian Statistics on Medicines 2016 1/41
Estonian Statistics on Medicines 2016 ATC code ATC group / Active substance (rout of admin.) Quantity sold Unit DDD Unit DDD/1000/ day A ALIMENTARY TRACT AND METABOLISM 167,8985 A01 STOMATOLOGICAL PREPARATIONS 0,0738 A01A STOMATOLOGICAL PREPARATIONS 0,0738 A01AB Antiinfectives and antiseptics for local oral treatment 0,0738 A01AB09 Miconazole (O) 7088 g 0,2 g 0,0738 A01AB12 Hexetidine (O) 1951200 ml A01AB81 Neomycin+ Benzocaine (dental) 30200 pieces A01AB82 Demeclocycline+ Triamcinolone (dental) 680 g A01AC Corticosteroids for local oral treatment A01AC81 Dexamethasone+ Thymol (dental) 3094 ml A01AD Other agents for local oral treatment A01AD80 Lidocaine+ Cetylpyridinium chloride (gingival) 227150 g A01AD81 Lidocaine+ Cetrimide (O) 30900 g A01AD82 Choline salicylate (O) 864720 pieces A01AD83 Lidocaine+ Chamomille extract (O) 370080 g A01AD90 Lidocaine+ Paraformaldehyde (dental) 405 g A02 DRUGS FOR ACID RELATED DISORDERS 47,1312 A02A ANTACIDS 1,0133 Combinations and complexes of aluminium, calcium and A02AD 1,0133 magnesium compounds A02AD81 Aluminium hydroxide+ Magnesium hydroxide (O) 811120 pieces 10 pieces 0,1689 A02AD81 Aluminium hydroxide+ Magnesium hydroxide (O) 3101974 ml 50 ml 0,1292 A02AD83 Calcium carbonate+ Magnesium carbonate (O) 3434232 pieces 10 pieces 0,7152 DRUGS FOR PEPTIC ULCER AND GASTRO- A02B 46,1179 OESOPHAGEAL REFLUX DISEASE (GORD) A02BA H2-receptor antagonists 2,3855 A02BA02 Ranitidine (O) 340327,5 g 0,3 g 2,3624 A02BA02 Ranitidine (P) 3318,25 g 0,3 g 0,0230 A02BC Proton pump inhibitors 43,7324 A02BC01 Omeprazole -

(12) Patent Application Publication (10) Pub. No.: US 2009/0292100 A1 Fiene Et Al
US 20090292100A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2009/0292100 A1 Fiene et al. (43) Pub. Date: Nov. 26, 2009 (54) PROCESS FOR PREPARING (86). PCT No.: PCT/EP07/57646 PENTAMETHYLENE 1.5-DIISOCYANATE S371 (c)(1), (75) Inventors: Martin Fiene, Niederkirchen (DE): (2), (4) Date: Jan. 9, 2009 (DE);Eckhard Wolfgang Stroefer, Siegel, Mannheim (30) Foreign ApplicationO O Priority Data Limburgerhof (DE); Stephan Aug. 1, 2006 (EP) .................................. O61182.56.4 Freyer, Neustadt (DE); Oskar Zelder, Speyer (DE); Gerhard Publication Classification Schulz, Bad Duerkheim (DE) (51) Int. Cl. Correspondence Address: CSG 18/00 (2006.01) OBLON, SPIVAK, MCCLELLAND MAIER & CD7C 263/2 (2006.01) NEUSTADT, L.L.P. CI2P I3/00 (2006.01) 194O DUKE STREET CD7C 263/10 (2006.01) ALEXANDRIA, VA 22314 (US) (52) U.S. Cl. ........... 528/85; 560/348; 435/128; 560/347; 560/355 (73) Assignee: BASFSE, LUDWIGSHAFEN (DE) (57) ABSTRACT (21) Appl. No.: 12/373,088 The present invention relates to a process for preparing pen tamethylene 1,5-diisocyanate, to pentamethylene 1,5-diiso (22) PCT Filed: Jul. 25, 2007 cyanate prepared in this way and to the use thereof. US 2009/0292100 A1 Nov. 26, 2009 PROCESS FOR PREPARING ene diisocyanates, especially pentamethylene 1,4-diisocyan PENTAMETHYLENE 1.5-DIISOCYANATE ate. Depending on its preparation, this proportion may be up to several % by weight. 0014. The pentamethylene 1,5-diisocyanate prepared in 0001. The present invention relates to a process for pre accordance with the invention has, in contrast, a proportion of paring pentamethylene 1,5-diisocyanate, to pentamethylene the branched pentamethylene diisocyanate isomers of in each 1.5-diisocyanate prepared in this way and to the use thereof. -

Antidote List
Antidote List Recommended minimum amounts for hospital pharmacies to stock Call the Maryland Poison Center for expert advice on the treatment of all poisonings and overdoses: 1-800-222-1222 Poisoning Minimum Stocking Antidote Indication Recommendations Oral: 120 grams Acetylcysteine Acetaminophen IV: 96 grams Crotaline snake Antivenin, snake (CroFab®) 12 vials envenomation 1 vial Black widow spider (Note: Merck limits distribution Antivenin, black widow spider envenomation to cases of confirmed bites with symptoms only) Organophosphate & Atropine carbamate insecticides, nerve 165 mg gases Calcium chloride Calcium channel blockers 10 grams 2.25 grams Calcium disodium EDTA Lead (Available direct from ASD Specialty Healthcare) Hydrofluoric acid 1 kg (powder) Calcium gluconate Calcium channel blockers IV: 30 grams 1 kit Cyanide Antidote Kit Cyanide (or Hydroxocobalamin, see below) Cyproheptadine (Periactin®) Serotonin syndrome 80 mg Neuroleptic malignant Dantrolene syndrome, stimulant-induced 720 mg hyperthermia Deferoxamine mesylate Iron 8 grams (Desferal®) Digoxin Immune FAB Digoxin 20 vials (Digibind®, DigiFab®) Dimercaprol (BAL) Heavy metals 1.5 grams 1 | P a g e Maryland Poison Center Antidote List – continued Poisoning Minimum Stocking Antidote Indication Recommendations DMSA (Succimer, Chemet®) Heavy metals 2000 mg Folic acid Methanol IV: 150 mg Flumazenil (Romazicon®) Benzodiazepines 10 mg Fomepizole (Antizol®) Ethylene glycol, methanol 12 grams Beta blockers, Glucagon 50 mg calcium channel blockers Hydroxocobalamin (Cyanokit®) Cyanide -

Protamine Sulfate Enhances Lipid-Mediated Gene Transfer
Gene Therapy (1997) 4, 961–968 1997 Stockton Press All rights reserved 0969-7128/97 $12.00 Protamine sulfate enhances lipid-mediated gene transfer FL Sorgi1,2, S Bhattacharya1 and L Huang1 1The Laboratory of Drug Targeting, Department of Pharmacology, The University of Pittsburgh School of Medicine, Pittsburgh, PA, USA A polycationic peptide, protamine sulfate, USP, has been USP as a condensation agent was found to be superior to shown to be able to condense plasmid DNA efficiently for poly-L-lysine as well as to various other types of protamine. delivery into several different types of cells in vitro by sev- These differences among various salt forms of protamine eral different types of cationic liposomes. The monovalent appear to be attributable to structural differences between cationic liposomal formulations (DC-Chol and lipofectin) the protamines and not due to differences in the net charge exhibited increased transfection activities comparable to of the molecule. The appearance of lysine residues within that seen with the multivalent cationic liposome formu- the protamine molecule correlate with a reduction in bind- lation, lipofectamine. This suggests that lipofectamine’s ing affinity to plasmid DNA, as well as an observed loss in superior in vitro activity arises from its ability to condense transfection-enhancing activity. This finding sheds light on DNA efficiently and that protamine’s primary role is that of the structural requirements of condensation agents for use a condensation agent, although it also possesses several in gene transfer protocols. Furthermore, protamine sulfate, amino acid sequences resembling that of a nuclear localiz- USP is an FDA-approved compound with a documented ation signal. -

Reducing the Use of Reversal Agents in a Community Hospital
4/21/2016 Objectives • Review the appropriate use of reversal agents Reducing the Use of Reversal Agents in a Community Hospital • Identify opportunities for reduction strategies of reversal agents Maria Paulina Duarte, PharmD PGY-1 Pharmacy Resident • Understand pharmacist’s role in impacting the Mercy Hospital, A Campus of use of reversal agents Plantation General Hospital Background Reversal Agents • Drug overdoses, whether accidental or intentional, • Naloxone constitute a significant source of increasing ▫ Reversal of opioids morbidity, mortality, and healthcare expenditure Examples: Morphine, hydromorphone, and fentanyl worldwide1,2 ▫ Side effects: Cardiac complications • Flumazenil • Hospitalized patients are susceptible to accidental ▫ Reversal of benzodiazepines drug overdoses, and immediate reversal may be 3 Examples: Lorazepam, clonazepam, and diazepam warranted to decrease harm ▫ Side effects: May result in seizures • Vitamin K • However, in some clinical settings, it has become ▫ Reversal of vitamin K antagonist common practice to use reversal agents and in some Example: Warfarin occasions the use of reversal agents might not be of ▫ Side effects: May cause warfarin resistance value for a drug overdose3,4 Reversal Agents Study’s Objective • Protamine ▫ Reversal of heparin Example: Unfractionated heparin and low molecular To assess the utilization of reversal agents weight heparin ▫ Side effects: cardiac and pulmonary complications and identify areas to potentially streamline their use • Prothrombin Complex Concentrate -

Discovery and Characterization of Blse, a Radical S- Adenosyl-L-Methionine Decarboxylase Involved in the Blasticidin S Biosynthetic Pathway
Discovery and Characterization of BlsE, a Radical S- Adenosyl-L-methionine Decarboxylase Involved in the Blasticidin S Biosynthetic Pathway Jun Feng1, Jun Wu1, Nan Dai2, Shuangjun Lin1, H. Howard Xu3, Zixin Deng1, Xinyi He1* 1 State Key Laboratory of Microbial Metabolism and School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai, China, 2 New England Biolabs, Inc., Research Department, Ipswich, Massachusetts, United States of America, 3 Department of Biological Sciences, California State University Los Angeles, Los Angeles, California, United States of America Abstract BlsE, a predicted radical S-adenosyl-L-methionine (SAM) protein, was anaerobically purified and reconstituted in vitro to study its function in the blasticidin S biosynthetic pathway. The putative role of BlsE was elucidated based on bioinformatics analysis, genetic inactivation and biochemical characterization. Biochemical results showed that BlsE is a SAM-dependent radical enzyme that utilizes cytosylglucuronic acid, the accumulated intermediate metabolite in blsE mutant, as substrate and catalyzes decarboxylation at the C5 position of the glucoside residue to yield cytosylarabinopyranose. Additionally, we report the purification and reconstitution of BlsE, characterization of its [4Fe–4S] cluster using UV-vis and electron paramagnetic resonance (EPR) spectroscopic analysis, and investigation of the ability of flavodoxin (Fld), flavodoxin 2+ reductase (Fpr) and NADPH to reduce the [4Fe–4S] cluster. Mutagenesis studies demonstrated that Cys31, Cys35, Cys38 in the C666C6MC motif and Gly73, Gly74, Glu75, Pro76 in the GGEP motif were crucial amino acids for BlsE activity while mutation of Met37 had little effect on its function. Our results indicate that BlsE represents a typical [4Fe–4S]-containing radical SAM enzyme and it catalyzes decarboxylation in blasticidin S biosynthesis. -

World Health Organization Model List of Essential Medicines, 21St List, 2019
World Health Organizatio n Model List of Essential Medicines 21st List 2019 World Health Organizatio n Model List of Essential Medicines 21st List 2019 WHO/MVP/EMP/IAU/2019.06 © World Health Organization 2019 Some rights reserved. This work is available under the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 IGO licence (CC BY-NC-SA 3.0 IGO; https://creativecommons.org/licenses/by-nc-sa/3.0/igo). Under the terms of this licence, you may copy, redistribute and adapt the work for non-commercial purposes, provided the work is appropriately cited, as indicated below. In any use of this work, there should be no suggestion that WHO endorses any specific organization, products or services. The use of the WHO logo is not permitted. If you adapt the work, then you must license your work under the same or equivalent Creative Commons licence. If you create a translation of this work, you should add the following disclaimer along with the suggested citation: “This translation was not created by the World Health Organization (WHO). WHO is not responsible for the content or accuracy of this translation. The original English edition shall be the binding and authentic edition”. Any mediation relating to disputes arising under the licence shall be conducted in accordance with the mediation rules of the World Intellectual Property Organization. Suggested citation. World Health Organization Model List of Essential Medicines, 21st List, 2019. Geneva: World Health Organization; 2019. Licence: CC BY-NC-SA 3.0 IGO. Cataloguing-in-Publication (CIP) data. CIP data are available at http://apps.who.int/iris. -

Phytohormones Regulate Accumulation of Osmolytes Under Abiotic Stress
biomolecules Review Phytohormones Regulate Accumulation of Osmolytes Under Abiotic Stress Anket Sharma 1,* , Babar Shahzad 2, Vinod Kumar 3 , Sukhmeen Kaur Kohli 4, Gagan Preet Singh Sidhu 5, Aditi Shreeya Bali 6, Neha Handa 7 , Dhriti Kapoor 7, Renu Bhardwaj 4 and Bingsong Zheng 1,* 1 State Key Laboratory of Subtropical Silviculture, Zhejiang A&F University, Hangzhou 311300, China 2 School of Land and Food, University of Tasmania, Hobart, Tasmania 7005, Australia 3 Department of Botany, DAV University, Sarmastpur, Jalandhar 144012, Punjab, India 4 Plant Stress Physiology Laboratory, Department of Botanical & Environmental Sciences, Guru Nanak Dev University, Amritsar 143005, India 5 Department of Environment Education, Government College of Commerce and Business Administration, Chandigarh 160047, India 6 Mehr Chand Mahajan D.A.V. College for Women, Chandigarh 160036, India 7 School of Bioengineering & Biosciences, Lovely Professional University, Phagwara 144411, India * Correspondence: [email protected] (A.S.); [email protected] (B.Z.); Tel.: +86-(0)-5716-373-0936 (B.Z.) Received: 13 June 2019; Accepted: 16 July 2019; Published: 17 July 2019 Abstract: Plants face a variety of abiotic stresses, which generate reactive oxygen species (ROS), and ultimately obstruct normal growth and development of plants. To prevent cellular damage caused by oxidative stress, plants accumulate certain compatible solutes known as osmolytes to safeguard the cellular machinery. The most common osmolytes that play crucial role in osmoregulation are proline, glycine-betaine, polyamines, and sugars. These compounds stabilize the osmotic differences between surroundings of cell and the cytosol. Besides, they also protect the plant cells from oxidative stress by inhibiting the production of harmful ROS like hydroxyl ions, superoxide ions, hydrogen peroxide, and other free radicals.