Symbol Low Grade High Grade GJA3 -2.71150382 1.68300234
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Different Genetic Mechanisms Mediate Spontaneous Versus UVR-Induced
RESEARCH ARTICLE Different genetic mechanisms mediate spontaneous versus UVR-induced malignant melanoma Blake Ferguson1, Herlina Y Handoko1, Pamela Mukhopadhyay1, Arash Chitsazan1, Lois Balmer2,3, Grant Morahan2, Graeme J Walker1†* 1Drug Discovery Group, QIMR Berghofer Medical Research Institute, Herston, Australia; 2Centre for Diabetes Research, Harry Perkins Institute of Medical Research, Perth, Australia; 3School of Medical and Health Sciences, Edith Cowan University, Joondalup, Australia Abstract Genetic variation conferring resistance and susceptibility to carcinogen-induced tumorigenesis is frequently studied in mice. We have now turned this idea to melanoma using the collaborative cross (CC), a resource of mouse strains designed to discover genes for complex diseases. We studied melanoma-prone transgenic progeny across seventy CC genetic backgrounds. We mapped a strong quantitative trait locus for rapid onset spontaneous melanoma onset to Prkdc, a gene involved in detection and repair of DNA damage. In contrast, rapid onset UVR-induced melanoma was linked to the ribosomal subunit gene Rrp15. Ribosome biogenesis was upregulated in skin shortly after UVR exposure. Mechanistically, variation in the ‘usual suspects’ by which UVR may exacerbate melanoma, defective DNA repair, melanocyte proliferation, or inflammatory cell infiltration, did not explain melanoma susceptibility or resistance across the CC. *For correspondence: [email protected] Instead, events occurring soon after exposure, such as dysregulation of ribosome function, which alters many aspects of cellular metabolism, may be important. † Present address: Experimental DOI: https://doi.org/10.7554/eLife.42424.001 Dermatology Group, The University of Queensland Diamantina Institute, Woolloongabba, Australia Introduction Competing interests: The Cutaneous malignant melanoma (MM) is well known to be associated with high levels of sun expo- authors declare that no sure. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

DULIP: a Dual Luminescence-Based Co-Immunoprecipitation Assay for Interactome Mapping in Mammalian Cells
Repository of the Max Delbrück Center for Molecular Medicine (MDC) in the Helmholtz Association http://edoc.mdc-berlin.de/14998 DULIP: A dual luminescence-based co-immunoprecipitation assay for interactome mapping in mammalian cells Trepte, P., Buntru, A., Klockmeier, K., Willmore, L., Arumughan, A., Secker, C., Zenkner, M., Brusendorf, L., Rau, K., Redel, A., Wanker, E.E. NOTICE: this is the author’s version of a work that was accepted for publication in the Journal of Molecular Biology. Changes resulting from the publishing process, such as peer review, editing, corrections, structural formatting, and other quality control mechanisms may not be reflected in this document. Changes may have been made to this work since it was submitted for publication. A definitive version was subsequently published in: Journal of Molecular Biology 2015 MMM DD ; 427(21): 3375-3388 doi: 10.1016/j.jmb.2015.08.003 Publisher: Elsevier © 2015, Elsevier. This work is licensed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/ or send a letter to Creative Commons, PO Box 1866, Mountain View, CA 94042, USA. DULIP: A DUAL LUMINESCENCE-BASED CO-IMMUNOPRECIPITATION ASSAY FOR INTERACTOME MAPPING IN MAMMALIAN CELLS Philipp Treptea, Alexander Buntrua#, Konrad Klockmeiera#, Lindsay Willmorea, Anup Arumughana, Christopher Seckera, Martina Zenknera, Lydia Brusendorfa, Kirstin Raua, Alexandra Redela and Erich E Wankera* a Neuroproteomics, Max Delbrueck Center for Molecular Medicine, Robert-Roessle- Straße 10, 13125 Berlin, Germany # Contributed equally * Corresponding author, E-mail address: [email protected], Telephone: +49- 30-9406-2157, Fax: +49-30-9406-2552 ABSTRACT Mapping of protein-protein interactions (PPIs) is critical for understanding protein function and complex biological processes. -

Supplementary Figure S4
18DCIS 18IDC Supplementary FigureS4 22DCIS 22IDC C D B A E (0.77) (0.78) 16DCIS 14DCIS 28DCIS 16IDC 28IDC (0.43) (0.49) 0 ADAMTS12 (p.E1469K) 14IDC ERBB2, LASP1,CDK12( CCNE1 ( NUTM2B SDHC,FCGR2B,PBX1,TPR( CD1D, B4GALT3, BCL9, FLG,NUP21OL,TPM3,TDRD10,RIT1,LMNA,PRCC,NTRK1 0 ADAMTS16 (p.E67K) (0.67) (0.89) (0.54) 0 ARHGEF38 (p.P179Hfs*29) 0 ATG9B (p.P823S) (0.68) (1.0) ARID5B, CCDC6 CCNE1, TSHZ3,CEP89 CREB3L2,TRIM24 BRAF, EGFR (7p11); 0 ABRACL (p.R35H) 0 CATSPER1 (p.P152H) 0 ADAMTS18 (p.Y799C) 19q12 0 CCDC88C (p.X1371_splice) (0) 0 ADRA1A (p.P327L) (10q22.3) 0 CCNF (p.D637N) −4 −2 −4 −2 0 AKAP4 (p.G454A) 0 CDYL (p.Y353Lfs*5) −4 −2 Log2 Ratio Log2 Ratio −4 −2 Log2 Ratio Log2 Ratio 0 2 4 0 2 4 0 ARID2 (p.R1068H) 0 COL27A1 (p.G646E) 0 2 4 0 2 4 2 EDRF1 (p.E521K) 0 ARPP21 (p.P791L) ) 0 DDX11 (p.E78K) 2 GPR101, p.A174V 0 ARPP21 (p.P791T) 0 DMGDH (p.W606C) 5 ANP32B, p.G237S 16IDC (Ploidy:2.01) 16DCIS (Ploidy:2.02) 14IDC (Ploidy:2.01) 14DCIS (Ploidy:2.9) -3 -2 -1 -3 -2 -1 -3 -2 -1 -3 -2 -1 -3 -2 -1 -3 -2 -1 Log Ratio Log Ratio Log Ratio Log Ratio 12DCIS 0 ASPM (p.S222T) Log Ratio Log Ratio 0 FMN2 (p.G941A) 20 1 2 3 2 0 1 2 3 2 ERBB3 (p.D297Y) 2 0 1 2 3 20 1 2 3 0 ATRX (p.L1276I) 20 1 2 3 2 0 1 2 3 0 GALNT18 (p.F92L) 2 MAPK4, p.H147Y 0 GALNTL6 (p.E236K) 5 C11orf1, p.Y53C (10q21.2); 0 ATRX (p.R1401W) PIK3CA, p.H1047R 28IDC (Ploidy:2.0) 28DCIS (Ploidy:2.0) 22IDC (Ploidy:3.7) 22DCIS (Ploidy:4.1) 18IDC (Ploidy:3.9) 18DCIS (Ploidy:2.3) 17q12 0 HCFC1 (p.S2025C) 2 LCMT1 (p.S34A) 0 ATXN7L2 (p.X453_splice) SPEN, p.P677Lfs*13 CBFB 1 2 3 4 5 6 7 8 9 10 11 -

Supplementary File 2A Revised
Supplementary file 2A. Differentially expressed genes in aldosteronomas compared to all other samples, ranked according to statistical significance. Missing values were not allowed in aldosteronomas, but to a maximum of five in the other samples. Acc UGCluster Name Symbol log Fold Change P - Value Adj. P-Value B R99527 Hs.8162 Hypothetical protein MGC39372 MGC39372 2,17 6,3E-09 5,1E-05 10,2 AA398335 Hs.10414 Kelch domain containing 8A KLHDC8A 2,26 1,2E-08 5,1E-05 9,56 AA441933 Hs.519075 Leiomodin 1 (smooth muscle) LMOD1 2,33 1,3E-08 5,1E-05 9,54 AA630120 Hs.78781 Vascular endothelial growth factor B VEGFB 1,24 1,1E-07 2,9E-04 7,59 R07846 Data not found 3,71 1,2E-07 2,9E-04 7,49 W92795 Hs.434386 Hypothetical protein LOC201229 LOC201229 1,55 2,0E-07 4,0E-04 7,03 AA454564 Hs.323396 Family with sequence similarity 54, member B FAM54B 1,25 3,0E-07 5,2E-04 6,65 AA775249 Hs.513633 G protein-coupled receptor 56 GPR56 -1,63 4,3E-07 6,4E-04 6,33 AA012822 Hs.713814 Oxysterol bining protein OSBP 1,35 5,3E-07 7,1E-04 6,14 R45592 Hs.655271 Regulating synaptic membrane exocytosis 2 RIMS2 2,51 5,9E-07 7,1E-04 6,04 AA282936 Hs.240 M-phase phosphoprotein 1 MPHOSPH -1,40 8,1E-07 8,9E-04 5,74 N34945 Hs.234898 Acetyl-Coenzyme A carboxylase beta ACACB 0,87 9,7E-07 9,8E-04 5,58 R07322 Hs.464137 Acyl-Coenzyme A oxidase 1, palmitoyl ACOX1 0,82 1,3E-06 1,2E-03 5,35 R77144 Hs.488835 Transmembrane protein 120A TMEM120A 1,55 1,7E-06 1,4E-03 5,07 H68542 Hs.420009 Transcribed locus 1,07 1,7E-06 1,4E-03 5,06 AA410184 Hs.696454 PBX/knotted 1 homeobox 2 PKNOX2 1,78 2,0E-06 -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Nucleolus: a Central Hub for Nuclear Functions Olga Iarovaia, Elizaveta Minina, Eugene Sheval, Daria Onichtchouk, Svetlana Dokudovskaya, Sergey Razin, Yegor Vassetzky
Nucleolus: A Central Hub for Nuclear Functions Olga Iarovaia, Elizaveta Minina, Eugene Sheval, Daria Onichtchouk, Svetlana Dokudovskaya, Sergey Razin, Yegor Vassetzky To cite this version: Olga Iarovaia, Elizaveta Minina, Eugene Sheval, Daria Onichtchouk, Svetlana Dokudovskaya, et al.. Nucleolus: A Central Hub for Nuclear Functions. Trends in Cell Biology, Elsevier, 2019, 29 (8), pp.647-659. 10.1016/j.tcb.2019.04.003. hal-02322927 HAL Id: hal-02322927 https://hal.archives-ouvertes.fr/hal-02322927 Submitted on 18 Nov 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Nucleolus: A Central Hub for Nuclear Functions Olga Iarovaia, Elizaveta Minina, Eugene Sheval, Daria Onichtchouk, Svetlana Dokudovskaya, Sergey Razin, Yegor Vassetzky To cite this version: Olga Iarovaia, Elizaveta Minina, Eugene Sheval, Daria Onichtchouk, Svetlana Dokudovskaya, et al.. Nucleolus: A Central Hub for Nuclear Functions. Trends in Cell Biology, Elsevier, 2019, 29 (8), pp.647-659. 10.1016/j.tcb.2019.04.003. hal-02322927 HAL Id: hal-02322927 https://hal.archives-ouvertes.fr/hal-02322927 Submitted on 18 Nov 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. -

Table S8. Positively Selected Genes (Psgs) Identified in Glyptosternoid and Yellowhead Catfish Lineages
Table S8. positively selected genes (PSGs) identified in glyptosternoid and yellowhead catfish lineages. Lineage Gene ID Gene name Gene description P-value Corrected P-value G. maculatum ENSDARG00000000001 slc35a5 solute carrier family 35, member A5 0 0 G. maculatum ENSDARG00000000656 psmb9a proteasome (prosome, macropain) subunit, beta type, 9a 0 0 aldo-keto reductase family 7, member A3 (aflatoxin aldehyde G. maculatum ENSDARG00000016649 akr7a3 0 0 reductase) G. maculatum ENSDARG00000017422 apmap adipocyte plasma membrane associated protein 0 0 G. maculatum ENSDARG00000003813 srp54 signal recognition particle 54 0 0 G. maculatum ENSDARG00000016173 cct3 chaperonin containing TCP1, subunit 3 (gamma) 0.012102523 0.049848505 G. maculatum ENSDARG00000018049 sf3b2 splicing factor 3b, subunit 2 0 0 G. maculatum ENSDARG00000004581 sel1l sel-1 suppressor of lin-12-like (C. elegans) 0.004079946 0.01759805 G. maculatum ENSDARG00000020344 slc2a8 solute carrier family 2 (facilitated glucose transporter), member 8 0.00E+00 0 G. maculatum ENSDARG00000011885 mrpl19 mitochondrial ribosomal protein L19 0 0 G. maculatum ENSDARG00000012640 cideb cell death-inducing DFFA-like effector b 0.00112672 0.005077819 G. maculatum ENSDARG00000003127 zgc:123105 zgc:123105 0 0 G. maculatum ENSDARG00000012929 eif2d eukaryotic translation initiation factor 2D 0.001049906 0.004752953 G. maculatum ENSDARG00000012947 SKA2 spindle and kinetochore associated complex subunit 2 0 0 G. maculatum ENSDARG00000015851 pnn pinin, desmosome associated protein 0 0 G. maculatum ENSDARG00000011418 sigmar1 sigma non-opioid intracellular receptor 1 0.00E+00 0 G. maculatum ENSDARG00000006926 btd biotinidase 0 0 G. maculatum ENSDARG00000012674 rpusd4 RNA pseudouridylate synthase domain containing 4 0.00E+00 0 G. maculatum ENSDARG00000017389 igfbp7 insulin-like growth factor binding protein 7 1.05E-02 0.043622349 G. -

Bioinformatics Analysis for the Identification of Differentially Expressed Genes and Related Signaling Pathways in H
Bioinformatics analysis for the identification of differentially expressed genes and related signaling pathways in H. pylori-CagA transfected gastric cancer cells Dingyu Chen*, Chao Li, Yan Zhao, Jianjiang Zhou, Qinrong Wang and Yuan Xie* Key Laboratory of Endemic and Ethnic Diseases , Ministry of Education, Guizhou Medical University, Guiyang, China * These authors contributed equally to this work. ABSTRACT Aim. Helicobacter pylori cytotoxin-associated protein A (CagA) is an important vir- ulence factor known to induce gastric cancer development. However, the cause and the underlying molecular events of CagA induction remain unclear. Here, we applied integrated bioinformatics to identify the key genes involved in the process of CagA- induced gastric epithelial cell inflammation and can ceration to comprehend the potential molecular mechanisms involved. Materials and Methods. AGS cells were transected with pcDNA3.1 and pcDNA3.1::CagA for 24 h. The transfected cells were subjected to transcriptome sequencing to obtain the expressed genes. Differentially expressed genes (DEG) with adjusted P value < 0.05, | logFC |> 2 were screened, and the R package was applied for gene ontology (GO) enrichment and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. The differential gene protein–protein interaction (PPI) network was constructed using the STRING Cytoscape application, which conducted visual analysis to create the key function networks and identify the key genes. Next, the Submitted 20 August 2020 Kaplan–Meier plotter survival analysis tool was employed to analyze the survival of the Accepted 11 March 2021 key genes derived from the PPI network. Further analysis of the key gene expressions Published 15 April 2021 in gastric cancer and normal tissues were performed based on The Cancer Genome Corresponding author Atlas (TCGA) database and RT-qPCR verification. -
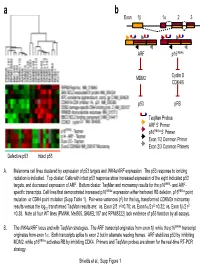
Taqman Probes ARF 5' Primer P16ink4a 5' Primer Exon 1/2
a b Exon 1β 1α 23 ARF p16INK4a MDM2 Cyclin D CDK4/6 p53 pRB TaqMan Probes ARF 5’ Primer p16INK4a 5’ Primer Exon 1/2 Common Primer Exon 2/3 Common Primers Defective p53 Intact p53 A. Melanoma cell lines clustered by expression of p53 targets and INK4a/ARF expression. The p53 response to ionizing radiation is indicated. Top cluster: Cells with intact p53 response show increased expression of the eight indicated p53 targets, and decreased expression of ARF. Bottom cluster: TaqMan and microarray results for the p16INK4- and ARF- specific transcripts. Cell lines that demonstrated increased p16INK4a expression either harbored RB deletion, p16INK4a point 2 mutation or CDK4 point mutation (Supp Table 1). Pair-wise variances (r ) for the log2 transformed CDKN2a microarray 2 2 2 results versus the log10 transformed TaqMan results are: vs. Exon 2/3 r =0.78; vs. Exon1α/2 r =0.32; vs. Exon 1β/2 r =0.38. Note all four WT lines (PMWK, Mel505, SKMEL187 and RPMI8322) lack evidence of p53 function by all assays. B. The INK4a/ARF locus and with TaqMan strategies. The ARF transcript originates from exon 1β while the p16INK4a transcript originates from exon 1α. Both transcripts splice to exon 2 but in alternate reading frames. ARF stabilizes p53 by inhibiting MDM2, while p16INK4a activates RB by inhibiting CDK4. Primers and TaqMan probes are shown for the real-time RT-PCR strategy. Shields et al., Supp Figure 1 SKMEL 28 U01 24h SKMEL WM2664 U01 48h WM2664 U01 24h 24 U01 48h SKMEL 24 U01 24h SKMEL 24 Untreated SKMEL 24 DMSO 48h SKMEL 24 DMSO 24h SKMEL WM2664 DMSO 24h WM2664 DMSO 48h WM2644 Untreated 28 Untreated SKMEL 28 DMSO 48h SKMEL 28 DMSO 24h SKMEL -3.00 -2.00 -1.00 0.00 1.00 2.00 3.00 relative to median expression Genes decreased by UO1 (863) HIF1A Hypoxia-inducible factor 1, alpha subunit NM_001530 RBBP8 Retinoblastoma binding protein 8 NM_002894 Homo sapiens, clone IMAGE:4337652, mRNA BC018676 EIF4EBP1 Eukaryotic translation initiation factor 4E binding protein EXOSC8 Exosome component 8 NM_181503 ENST00000321524 MCM7 MCM7 minichromosome maintenance deficient 7 (S. -
(A) Up-Regulated Genes in HCC827-GR-High2 Compared to Parental HCC827
Table S2. Results of expression profiling analysis (A) Up-regulated genes in HCC827-GR-high2 compared to parental HCC827 Fold Fold Fold Unique ID Symbol Unique ID Symbol Unique ID Symbol change* change* change* ILMN_1709348 ALDH1A1 577.587 ILMN_2310814 MAPT 13.003 ILMN_1741017 PIP4K2B 7.331 ILMN_1651354 SPP1 441.316 ILMN_1748650 MRPL45 12.988 ILMN_3237623 RNY1 7.297 ILMN_1701831 GSTA1 260.591 ILMN_1755897 UGT2B7 12.629 ILMN_1734897 SLC4A4 7.285 ILMN_1658835 CAV2 12.357 ILMN_1746359 RERG 7.280 ILMN_2094875 ABCB1 183.050 ILMN_1678939 VNN2 11.935 ILMN_1671337 SLC2A5 7.257 ILMN_3251540 GSTA2 145.982 ILMN_1729905 GAL3ST1 11.910 ILMN_1691606 LYG2 7.254 ILMN_2062468 IGFBP7 127.721 ILMN_1672536 FBLN1 11.716 ILMN_1785646 PMP22 7.246 ILMN_1795190 CLDN2 111.439 ILMN_1796339 PLEKHA2 11.631 ILMN_1737387 LOC728441 7.207 ILMN_1782937 LOC647169 98.612 ILMN_1676563 HTRA1 11.592 ILMN_1684401 FMO1 7.117 ILMN_1754247 SLC3A1 81.001 ILMN_3263423 LOC100129027 11.346 ILMN_1687035 ADAMTSL4 7.098 ILMN_1662795 CA2 79.581 ILMN_1694898 LOC653857 10.906 ILMN_2153572 MAGEA3 7.086 ILMN_2168747 GSTA2 66.250 ILMN_2404625 LAT 10.560 ILMN_1784283 USH1C 7.079 ILMN_1764228 DAB2 64.709 ILMN_1666546 DUSP14 10.375 ILMN_1731374 CPE 7.046 ILMN_1675797 EPDR1 63.605 ILMN_1764571 ARHGAP23 10.299 ILMN_1765446 EMP3 6.933 ILMN_1708341 PDZK1 59.714 ILMN_3200140 LOC645638 10.284 ILMN_1754002 IL1F8 6.863 ILMN_1713529 SEMA6A 52.575 ILMN_3244343 SNORA21 10.171 ILMN_1878007 FUT9 6.835 ILMN_1708391 NR1H4 43.218 ILMN_1671489 PC 10.075 ILMN_1699208 NAP1L1 6.763 ILMN_2412336 AKR1C2 42.826 ILMN_2404688 -

Supplementary Table 1
Supplementary Table 1. 492 genes are unique to 0 h post-heat timepoint. The name, p-value, fold change, location and family of each gene are indicated. Genes were filtered for an absolute value log2 ration 1.5 and a significance value of p ≤ 0.05. Symbol p-value Log Gene Name Location Family Ratio ABCA13 1.87E-02 3.292 ATP-binding cassette, sub-family unknown transporter A (ABC1), member 13 ABCB1 1.93E-02 −1.819 ATP-binding cassette, sub-family Plasma transporter B (MDR/TAP), member 1 Membrane ABCC3 2.83E-02 2.016 ATP-binding cassette, sub-family Plasma transporter C (CFTR/MRP), member 3 Membrane ABHD6 7.79E-03 −2.717 abhydrolase domain containing 6 Cytoplasm enzyme ACAT1 4.10E-02 3.009 acetyl-CoA acetyltransferase 1 Cytoplasm enzyme ACBD4 2.66E-03 1.722 acyl-CoA binding domain unknown other containing 4 ACSL5 1.86E-02 −2.876 acyl-CoA synthetase long-chain Cytoplasm enzyme family member 5 ADAM23 3.33E-02 −3.008 ADAM metallopeptidase domain Plasma peptidase 23 Membrane ADAM29 5.58E-03 3.463 ADAM metallopeptidase domain Plasma peptidase 29 Membrane ADAMTS17 2.67E-04 3.051 ADAM metallopeptidase with Extracellular other thrombospondin type 1 motif, 17 Space ADCYAP1R1 1.20E-02 1.848 adenylate cyclase activating Plasma G-protein polypeptide 1 (pituitary) receptor Membrane coupled type I receptor ADH6 (includes 4.02E-02 −1.845 alcohol dehydrogenase 6 (class Cytoplasm enzyme EG:130) V) AHSA2 1.54E-04 −1.6 AHA1, activator of heat shock unknown other 90kDa protein ATPase homolog 2 (yeast) AK5 3.32E-02 1.658 adenylate kinase 5 Cytoplasm kinase AK7