The Transcription Factor Rreb1 Regulates Epithelial Architecture, Invasiveness, and Vasculogenesis in Early Mouse Embryos
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

1 Evidence for Gliadin Antibodies As Causative Agents in Schizophrenia
1 Evidence for gliadin antibodies as causative agents in schizophrenia. C.J.Carter PolygenicPathways, 20 Upper Maze Hill, Saint-Leonard’s on Sea, East Sussex, TN37 0LG [email protected] Tel: 0044 (0)1424 422201 I have no fax Abstract Antibodies to gliadin, a component of gluten, have frequently been reported in schizophrenia patients, and in some cases remission has been noted following the instigation of a gluten free diet. Gliadin is a highly immunogenic protein, and B cell epitopes along its entire immunogenic length are homologous to the products of numerous proteins relevant to schizophrenia (p = 0.012 to 3e-25). These include members of the DISC1 interactome, of glutamate, dopamine and neuregulin signalling networks, and of pathways involved in plasticity, dendritic growth or myelination. Antibodies to gliadin are likely to cross react with these key proteins, as has already been observed with synapsin 1 and calreticulin. Gliadin may thus be a causative agent in schizophrenia, under certain genetic and immunological conditions, producing its effects via antibody mediated knockdown of multiple proteins relevant to the disease process. Because of such homology, an autoimmune response may be sustained by the human antigens that resemble gliadin itself, a scenario supported by many reports of immune activation both in the brain and in lymphocytes in schizophrenia. Gluten free diets and removal of such antibodies may be of therapeutic benefit in certain cases of schizophrenia. 2 Introduction A number of studies from China, Norway, and the USA have reported the presence of gliadin antibodies in schizophrenia 1-5. Gliadin is a component of gluten, intolerance to which is implicated in coeliac disease 6. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Transcriptional Control of Tissue-Resident Memory T Cell Generation
Transcriptional control of tissue-resident memory T cell generation Filip Cvetkovski Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Graduate School of Arts and Sciences COLUMBIA UNIVERSITY 2019 © 2019 Filip Cvetkovski All rights reserved ABSTRACT Transcriptional control of tissue-resident memory T cell generation Filip Cvetkovski Tissue-resident memory T cells (TRM) are a non-circulating subset of memory that are maintained at sites of pathogen entry and mediate optimal protection against reinfection. Lung TRM can be generated in response to respiratory infection or vaccination, however, the molecular pathways involved in CD4+TRM establishment have not been defined. Here, we performed transcriptional profiling of influenza-specific lung CD4+TRM following influenza infection to identify pathways implicated in CD4+TRM generation and homeostasis. Lung CD4+TRM displayed a unique transcriptional profile distinct from spleen memory, including up-regulation of a gene network induced by the transcription factor IRF4, a known regulator of effector T cell differentiation. In addition, the gene expression profile of lung CD4+TRM was enriched in gene sets previously described in tissue-resident regulatory T cells. Up-regulation of immunomodulatory molecules such as CTLA-4, PD-1, and ICOS, suggested a potential regulatory role for CD4+TRM in tissues. Using loss-of-function genetic experiments in mice, we demonstrate that IRF4 is required for the generation of lung-localized pathogen-specific effector CD4+T cells during acute influenza infection. Influenza-specific IRF4−/− T cells failed to fully express CD44, and maintained high levels of CD62L compared to wild type, suggesting a defect in complete differentiation into lung-tropic effector T cells. -

The Characterization and Role of RREB1 in Human Bladder Cancer
The Characterization and Role of RREB1 in Human Bladder Cancer Matthew David Nitz Sebring, Fl B.S. Wheaton College, 2003 M.S ., University of Virginia, 2009 A Dissertation presented to the Graduate Faculty of the University of Virginia in Cand idacy for the Degree of Doctor of Philosophy Department of Molecular Physiology and Biological Physics University of Virginia August, 2011 I Abstract RREB1 is a C2H2 zinc-finger protein that has previously been associated with malignancy but little is known about its expression, composition of splice variants, or role in cancer phenotypes. Total RREB1 expression decreased in human bladder cancer tumors compared to paired normal tissue. Rigorous comparison of commercial RREB1 antibodies allowed for the identification of RREB1 splice variants in the cytoplasm and nucleus on western blot. Five RREB1 splice variants were identified and designated: RREB1α, RREB1β, RREB1γ, RREB1δ, RREB1ε. Immunohistochemistry of RREB1 in a bladder cancer tissue microarray demonstrated that RREB1 expression positively correlates with the squamous cell histology. Transient depletion of RREB1 in bladder cancer cell lines (UMUC3 and KU7) decreased cell growth in vitro and similar results were seen in prostate cancer cell lines (LNCAP and PC3). siRNA duplexes that target RREB1 isoforms showed RREB1α and RREB1β, but not RREB1β alone, were necessary for in vitro UMUC3 cell growth and in vivo subcutaneous tumor growth. RREB1 depletion was found to decrease the expression of tumor associated protein CD24. Cloning of the CD24 promoter revealed a hypoxia response element, which led to the discovery of hypoxia induced CD24 RNA expression and promoter activity. RREB1 depletion decreased the expression of two additional hypoxia responsive genes (VEGFA and IGFBP3) without affecting the mRNA stability. -
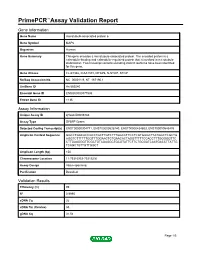
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name microtubule-associated protein 6 Gene Symbol MAP6 Organism Human Gene Summary This gene encodes a microtubule-associated protein. The encoded protein is a calmodulin-binding and calmodulin-regulated protein that is involved in microtubule stabilization. Two transcript variants encoding distinct isoforms have been identified for this gene. Gene Aliases FLJ41346, KIAA1878, MTAP6, N-STOP, STOP RefSeq Accession No. NC_000011.9, NT_167190.1 UniGene ID Hs.585540 Ensembl Gene ID ENSG00000171533 Entrez Gene ID 4135 Assay Information Unique Assay ID qHsaCID0006783 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000304771, ENST00000526740, ENST00000434603, ENST00000545476 Amplicon Context Sequence GGCCTGACACCGCCTGCTTGTCTTTGGCCTTCCTCGTGGGCTTATGGCTCGCTG AGGTCTTTTTTGGTTTGGAACTCTGAACACTAGGTTTTTCCACCTTTGGGGGTTC CTTGAAGGGTTCGCTGTAGAGGCTGCGTATTCTTCTGCGATCAATGACCTTATTG TCAGCTGTTGTTGGCT Amplicon Length (bp) 150 Chromosome Location 11:75316953-75319236 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 99 R2 0.9995 cDNA Cq 26 cDNA Tm (Celsius) 85 gDNA Cq 31.54 Page 1/5 PrimePCR™Assay Validation Report Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 2/5 PrimePCR™Assay Validation Report MAP6, Human Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve analysis of above amplification Standard Curve Standard curve generated using 20 million copies of template diluted 10-fold to 20 copies Page 3/5 PrimePCR™Assay Validation Report Products used to generate validation data Real-Time PCR Instrument CFX384 Real-Time PCR Detection System Reverse Transcription Reagent iScript™ Advanced cDNA Synthesis Kit for RT-qPCR Real-Time PCR Supermix SsoAdvanced™ SYBR® Green Supermix Experimental Sample qPCR Human Reference Total RNA Data Interpretation Unique Assay ID This is a unique identifier that can be used to identify the assay in the literature and online. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Ahn Supp. Fig. 1 AB 1.5 ARRDC4 1.5 ARRDC4 * * * 1.0 1.0
Ahn_Supp. Fig. 1 AB 1.5 ARRDC4 1.5 ARRDC4 * * * 1.0 1.0 * * 0.5 * 0.5 * * * Relative mRNA levels mRNA Relative Relative mRNA levels mRNA Relative 0.0 0.0 1.5 MLXIP (MondoA) 1.5 MLXIP (MondoA) 1.0 1.0 0.5 0.5 Relative mRNA levels mRNA Relative Relative mRNA levels mRNA Relative 0.0 0.0 MondoA MondoA 0124824 Starvation (6h) -++++++ Glucose Starvation (h) Refeeding (h) --0.51248 C 1.5 ARRDC4 1.5 MLXIP (MondoA) † Con # KD 1.0 1.0 0.5 0.5 * * * * Relative mRNA levels mRNA Relative Relative mRNA levels mRNA Relative * * 0.0 0.0 BasalStarvation Refeeding BasalStarvation Refeeding MondoA Con + + - - + + - - + + - - KD - - + + - - + + - - + + BasalStarvation Refeeding Supplemental Figure 1. Glucose-mediated regulation of ARRDC4 is dependent on MondoA in human skeletal myotubes. (A) (top) ARRDC4 and MLXIP (MondoA) mRNA levels were determined by qRT-PCR in human skeletal myotubes following deprivation of glucose at the indicated time (n=4). (bottom) Representative Western blot analysis of MondoA demonstrating the effect of glucose deprivation. *p<0.05 vs. 0h. (B) (top) ARRDC4 and MLXIP (MondoA) expression in human myotubes following a 6h glucose removal and refeeding at the times indicated (n=4). (bottom) Corresponding Western blot analysis. *p<0.05 vs Starvation 6h. (C) (top) Expression of ARRDC4 and MLXIP in human myotubes following deprivation and refeeding of glucose in the absence or presence of siRNA-mediated MondoA KD (n=4). (bottom) Corresponding Western blot analysis. *p<0.05 vs siControl. # p<0.05. § p<0.05. The data represents mean ± SD. All statistical significance determined by one-way ANOVA with Tukey multiple comparison post-hoc test. -

Whole Exome Sequencing in Families at High Risk for Hodgkin Lymphoma: Identification of a Predisposing Mutation in the KDR Gene
Hodgkin Lymphoma SUPPLEMENTARY APPENDIX Whole exome sequencing in families at high risk for Hodgkin lymphoma: identification of a predisposing mutation in the KDR gene Melissa Rotunno, 1 Mary L. McMaster, 1 Joseph Boland, 2 Sara Bass, 2 Xijun Zhang, 2 Laurie Burdett, 2 Belynda Hicks, 2 Sarangan Ravichandran, 3 Brian T. Luke, 3 Meredith Yeager, 2 Laura Fontaine, 4 Paula L. Hyland, 1 Alisa M. Goldstein, 1 NCI DCEG Cancer Sequencing Working Group, NCI DCEG Cancer Genomics Research Laboratory, Stephen J. Chanock, 5 Neil E. Caporaso, 1 Margaret A. Tucker, 6 and Lynn R. Goldin 1 1Genetic Epidemiology Branch, Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, Bethesda, MD; 2Cancer Genomics Research Laboratory, Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, Bethesda, MD; 3Ad - vanced Biomedical Computing Center, Leidos Biomedical Research Inc.; Frederick National Laboratory for Cancer Research, Frederick, MD; 4Westat, Inc., Rockville MD; 5Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, Bethesda, MD; and 6Human Genetics Program, Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, Bethesda, MD, USA ©2016 Ferrata Storti Foundation. This is an open-access paper. doi:10.3324/haematol.2015.135475 Received: August 19, 2015. Accepted: January 7, 2016. Pre-published: June 13, 2016. Correspondence: [email protected] Supplemental Author Information: NCI DCEG Cancer Sequencing Working Group: Mark H. Greene, Allan Hildesheim, Nan Hu, Maria Theresa Landi, Jennifer Loud, Phuong Mai, Lisa Mirabello, Lindsay Morton, Dilys Parry, Anand Pathak, Douglas R. Stewart, Philip R. Taylor, Geoffrey S. Tobias, Xiaohong R. Yang, Guoqin Yu NCI DCEG Cancer Genomics Research Laboratory: Salma Chowdhury, Michael Cullen, Casey Dagnall, Herbert Higson, Amy A. -

Microtubule Organization and Microtubule- Associated Proteins (Maps)
Chapter 3 Microtubule Organization and Microtubule- Associated Proteins (MAPs) Elena Tortosa, Lukas C. Kapitein, and Casper C. Hoogenraad Abstract Dendrites have a unique microtubule organization. In vertebrates, den- dritic microtubules are organized in antiparallel bundles, oriented with their plus ends either pointing away or toward the soma. The mixed microtubule arrays control intracellular trafficking and local signaling pathways, and are essential for dendrite development and function. The organization of microtubule arrays largely depends on the combined function of different microtubule regulatory factors or generally named microtubule-associated proteins (MAPs). Classical MAPs, also called structural MAPs, were identified more than 20 years ago based on their ability to bind to and copurify with microtubules. Most classical MAPs bind along the microtubule lattice and regulate microtubule polymerization, bundling, and stabilization. Recent evidences suggest that classical MAPs also guide motor protein transport, interact with the actin cytoskeleton, and act in various neuronal signaling networks. Here, we give an overview of microtubule organization in dendrites and the role of classical MAPs in dendrite development, dendritic spine formation, and synaptic plasticity. Keywords Neuron • Dendrite • Cytoskeleton • Microtubule • Microtubule- associated protein • MAP1 • MAP2 • MAP4 • MAP6 • MAP7 • MAP9 • Tau 3.1 Introduction Microtubules (MTs) are cytoskeletal structures that play essential roles in all eukaryotic cells. MTs are important not only during cell division but also in non-dividing cells, where they are critical structures in numerous cellular processes such as cell motility, migration, differentiation, intracellular transport and organelle positioning. MTs are composed of two proteins, α- and β-tubulin, that form heterodimers and organize themselves in a head-to-tail manner. -

Transcriptomic Characterisation of the Molecular Mechanisms Induced by Rgma During Skeletal Muscle Hyperplasia and Hypertrophy
Transcriptomic Characterisation of the Molecular Mechanisms Induced by RGMa During Skeletal Muscle Hyperplasia and Hypertrophy Aline Gonçalves Lio Copola Universidade Federal de Minas Gerais Íria Gabriela Dias dos Santos Universidade Federal de Minas Gerais Luiz Lehmann Coutinho Universidade de São Paulo Luiz Eduardo Vieira Del Bem Universidade Federal de Minas Gerais Paulo Henrique de Almeida Campos Junior Federal University of São João del-Rei Júlia Meireles Nogueira Universidade Federal de Minas Gerais Alinne do Carmo Costa Universidade Federal de Minas Gerais Gerluza Aparecida Borges Silva Universidade Federal de Minas Gerais Erika Cristina Jorge ( [email protected] ) Universidade Federal de Minas Gerais Research Article Keywords: Axon Guidance, myogenesis, hypertrophy, hyperplasia, skeletal muscle differentiation, transcriptomic analysis Posted Date: June 29th, 2021 DOI: https://doi.org/10.21203/rs.3.rs-646954/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/25 Abstract Background: The repulsive guidance molecule a (RGMa) is a GPI-anchor axon guidance molecule rst found to play important roles during neuronal development. RGMa expression patterns and signalling pathways via Neogenin and/or as BMP coreceptors indicated that this axon guidance molecule could also be working in other processes and diseases, including during myogenesis. Previous works have consistently shown that RGMa is expressed in skeletal muscle cells and that its overexpression induces both nuclei accretion and hypertrophy in muscle cell lineages. However, the cellular components and molecular mechanisms induced by RGMa during the differentiation of skeletal muscle cells are poorly understood. In this work, the global transcription expression prole of RGMa-treated C2C12 myoblasts during the differentiation stage, obtained by RNA- seq, were reported. -

In This Table Protein Name, Uniprot Code, Gene Name P-Value
Supplementary Table S1: In this table protein name, uniprot code, gene name p-value and Fold change (FC) for each comparison are shown, for 299 of the 301 significantly regulated proteins found in both comparisons (p-value<0.01, fold change (FC) >+/-0.37) ALS versus control and FTLD-U versus control. Two uncharacterized proteins have been excluded from this list Protein name Uniprot Gene name p value FC FTLD-U p value FC ALS FTLD-U ALS Cytochrome b-c1 complex P14927 UQCRB 1.534E-03 -1.591E+00 6.005E-04 -1.639E+00 subunit 7 NADH dehydrogenase O95182 NDUFA7 4.127E-04 -9.471E-01 3.467E-05 -1.643E+00 [ubiquinone] 1 alpha subcomplex subunit 7 NADH dehydrogenase O43678 NDUFA2 3.230E-04 -9.145E-01 2.113E-04 -1.450E+00 [ubiquinone] 1 alpha subcomplex subunit 2 NADH dehydrogenase O43920 NDUFS5 1.769E-04 -8.829E-01 3.235E-05 -1.007E+00 [ubiquinone] iron-sulfur protein 5 ARF GTPase-activating A0A0C4DGN6 GIT1 1.306E-03 -8.810E-01 1.115E-03 -7.228E-01 protein GIT1 Methylglutaconyl-CoA Q13825 AUH 6.097E-04 -7.666E-01 5.619E-06 -1.178E+00 hydratase, mitochondrial ADP/ATP translocase 1 P12235 SLC25A4 6.068E-03 -6.095E-01 3.595E-04 -1.011E+00 MIC J3QTA6 CHCHD6 1.090E-04 -5.913E-01 2.124E-03 -5.948E-01 MIC J3QTA6 CHCHD6 1.090E-04 -5.913E-01 2.124E-03 -5.948E-01 Protein kinase C and casein Q9BY11 PACSIN1 3.837E-03 -5.863E-01 3.680E-06 -1.824E+00 kinase substrate in neurons protein 1 Tubulin polymerization- O94811 TPPP 6.466E-03 -5.755E-01 6.943E-06 -1.169E+00 promoting protein MIC C9JRZ6 CHCHD3 2.912E-02 -6.187E-01 2.195E-03 -9.781E-01 Mitochondrial 2- -

Mutational Mechanisms That Activate Wnt Signaling and Predict Outcomes in Colorectal Cancer Patients William Hankey1, Michael A
Published OnlineFirst December 6, 2017; DOI: 10.1158/0008-5472.CAN-17-1357 Cancer Genome and Epigenome Research Mutational Mechanisms That Activate Wnt Signaling and Predict Outcomes in Colorectal Cancer Patients William Hankey1, Michael A. McIlhatton1, Kenechi Ebede2, Brian Kennedy3, Baris Hancioglu3, Jie Zhang4, Guy N. Brock3, Kun Huang4, and Joanna Groden1 Abstract APC biallelic loss-of-function mutations are the most prevalent also exhibiting unique changes in pathways related to prolifera- genetic changes in colorectal tumors, but it is unknown whether tion, cytoskeletal organization, and apoptosis. Apc-mutant ade- these mutations phenocopy gain-of-function mutations in the nomas were characterized by increased expression of the glial CTNNB1 gene encoding b-catenin that also activate canonical nexin Serpine2, the human ortholog, which was increased in WNT signaling. Here we demonstrate that these two mutational advanced human colorectal tumors. Our results support the mechanisms are not equivalent. Furthermore, we show how hypothesis that APC-mutant colorectal tumors are transcription- differences in gene expression produced by these different ally distinct from APC-wild-type colorectal tumors with canonical mechanisms can stratify outcomes in more advanced human WNT signaling activated by other mechanisms, with possible colorectal cancers. Gene expression profiling in Apc-mutant and implications for stratification and prognosis. Ctnnb1-mutant mouse colon adenomas identified candidate Significance: These findings suggest that colon adenomas genes for subsequent evaluation of human TCGA (The Cancer driven by APC mutations are distinct from those driven by WNT Genome Atlas) data for colorectal cancer outcomes. Transcrip- gain-of-function mutations, with implications for identifying tional patterns exhibited evidence of activated canonical Wnt at-risk patients with advanced disease based on gene expression signaling in both types of adenomas, with Apc-mutant adenomas patterns.