Mutational Mechanisms That Activate Wnt Signaling and Predict Outcomes in Colorectal Cancer Patients William Hankey1, Michael A
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

1 Evidence for Gliadin Antibodies As Causative Agents in Schizophrenia
1 Evidence for gliadin antibodies as causative agents in schizophrenia. C.J.Carter PolygenicPathways, 20 Upper Maze Hill, Saint-Leonard’s on Sea, East Sussex, TN37 0LG [email protected] Tel: 0044 (0)1424 422201 I have no fax Abstract Antibodies to gliadin, a component of gluten, have frequently been reported in schizophrenia patients, and in some cases remission has been noted following the instigation of a gluten free diet. Gliadin is a highly immunogenic protein, and B cell epitopes along its entire immunogenic length are homologous to the products of numerous proteins relevant to schizophrenia (p = 0.012 to 3e-25). These include members of the DISC1 interactome, of glutamate, dopamine and neuregulin signalling networks, and of pathways involved in plasticity, dendritic growth or myelination. Antibodies to gliadin are likely to cross react with these key proteins, as has already been observed with synapsin 1 and calreticulin. Gliadin may thus be a causative agent in schizophrenia, under certain genetic and immunological conditions, producing its effects via antibody mediated knockdown of multiple proteins relevant to the disease process. Because of such homology, an autoimmune response may be sustained by the human antigens that resemble gliadin itself, a scenario supported by many reports of immune activation both in the brain and in lymphocytes in schizophrenia. Gluten free diets and removal of such antibodies may be of therapeutic benefit in certain cases of schizophrenia. 2 Introduction A number of studies from China, Norway, and the USA have reported the presence of gliadin antibodies in schizophrenia 1-5. Gliadin is a component of gluten, intolerance to which is implicated in coeliac disease 6. -

Genome-Wide Screen of Otosclerosis in Population Biobanks
medRxiv preprint doi: https://doi.org/10.1101/2020.11.15.20227868; this version posted November 16, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. It is made available under a CC-BY-NC-ND 4.0 International license . 1 Genome-wide Screen of Otosclerosis in 2 Population Biobanks: 18 Loci and Shared 3 Heritability with Skeletal Structure 4 Joel T. Rämö1, Tuomo Kiiskinen1, Juha Karjalainen1,2,3,4, Kristi Krebs5, Mitja Kurki1,2,3,4, Aki S. 5 Havulinna6, Eija Hämäläinen1, Paavo Häppölä1, Heidi Hautakangas1, FinnGen, Konrad J. 6 Karczewski1,2,3,4, Masahiro Kanai1,2,3,4, Reedik Mägi5, Priit Palta1,5, Tõnu Esko5, Andres Metspalu5, 7 Matti Pirinen1,7,8, Samuli Ripatti1,2,7, Lili Milani5, Antti Mäkitie9, Mark J. Daly1,2,3,4,10, and Aarno 8 Palotie1,2,3,4 9 1. Institute for Molecular Medicine Finland (FIMM), Helsinki Institute of Life Science (HiLIFE), University of 10 Helsinki, Helsinki, Finland 11 2. Program in Medical and Population Genetics, Broad Institute of Harvard and MIT, Cambridge, 12 Massachusetts, USA 13 3. Stanley Center for Psychiatric Research, Broad Institute of Harvard and MIT, Cambridge, Massachusetts, 14 USA 15 4. Analytic and Translational Genetics Unit, Massachusetts General Hospital, Boston, Massachusetts, USA 16 5. Estonian Genome Center, University of Tartu, Tartu, Estonia, Institute of Molecular and Cell Biology, 17 University of Tartu, Tartu, Estonia 18 6. Finnish Institute for Health and Welfare, Helsinki, Finland 19 7. Department of Public Health, Clinicum, Faculty of Medicine, University of Helsinki, Helsinki, Finland 20 8. -
Primepcr™Assay Validation Report
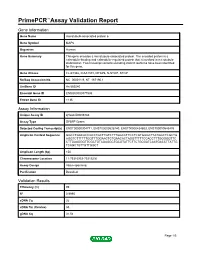
PrimePCR™Assay Validation Report Gene Information Gene Name microtubule-associated protein 6 Gene Symbol MAP6 Organism Human Gene Summary This gene encodes a microtubule-associated protein. The encoded protein is a calmodulin-binding and calmodulin-regulated protein that is involved in microtubule stabilization. Two transcript variants encoding distinct isoforms have been identified for this gene. Gene Aliases FLJ41346, KIAA1878, MTAP6, N-STOP, STOP RefSeq Accession No. NC_000011.9, NT_167190.1 UniGene ID Hs.585540 Ensembl Gene ID ENSG00000171533 Entrez Gene ID 4135 Assay Information Unique Assay ID qHsaCID0006783 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000304771, ENST00000526740, ENST00000434603, ENST00000545476 Amplicon Context Sequence GGCCTGACACCGCCTGCTTGTCTTTGGCCTTCCTCGTGGGCTTATGGCTCGCTG AGGTCTTTTTTGGTTTGGAACTCTGAACACTAGGTTTTTCCACCTTTGGGGGTTC CTTGAAGGGTTCGCTGTAGAGGCTGCGTATTCTTCTGCGATCAATGACCTTATTG TCAGCTGTTGTTGGCT Amplicon Length (bp) 150 Chromosome Location 11:75316953-75319236 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 99 R2 0.9995 cDNA Cq 26 cDNA Tm (Celsius) 85 gDNA Cq 31.54 Page 1/5 PrimePCR™Assay Validation Report Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 2/5 PrimePCR™Assay Validation Report MAP6, Human Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve analysis of above amplification Standard Curve Standard curve generated using 20 million copies of template diluted 10-fold to 20 copies Page 3/5 PrimePCR™Assay Validation Report Products used to generate validation data Real-Time PCR Instrument CFX384 Real-Time PCR Detection System Reverse Transcription Reagent iScript™ Advanced cDNA Synthesis Kit for RT-qPCR Real-Time PCR Supermix SsoAdvanced™ SYBR® Green Supermix Experimental Sample qPCR Human Reference Total RNA Data Interpretation Unique Assay ID This is a unique identifier that can be used to identify the assay in the literature and online. -

Nuclear Envelope Laminopathies: Evidence for Developmentally Inappropriate Nuclear Envelope-Chromatin Associations
Nuclear Envelope Laminopathies: Evidence for Developmentally Inappropriate Nuclear Envelope-Chromatin Associations by Jelena Perovanovic M.S. in Molecular Biology and Physiology, September 2009, University of Belgrade M.Phil. in Molecular Medicine, August 2013, The George Washington University A Dissertation submitted to The Faculty of The Columbian College of Arts and Sciences of The George Washington University in partial fulfillment of the requirements for the degree of Doctor of Philosophy August 31, 2015 Dissertation directed by Eric P. Hoffman Professor of Integrative Systems Biology The Columbian College of Arts and Sciences of The George Washington University certifies that Jelena Perovanovic has passed the Final Examination for the degree of Doctor of Philosophy as of May 5, 2015. This is the final and approved form of the dissertation. Nuclear Envelope Laminopathies: Evidence for Developmentally Inappropriate Nuclear Envelope-Chromatin Associations Jelena Perovanovic Dissertation Research Committee: Eric P. Hoffman, Professor of Integrative Systems Biology, Dissertation Director Anamaris Colberg-Poley, Professor of Integrative Systems Biology, Committee Member Robert J. Freishtat, Associate Professor of Pediatrics, Committee Member Vittorio Sartorelli, Senior Investigator, National Institutes of Health, Committee Member ii © Copyright 2015 by Jelena Perovanovic All rights reserved iii Acknowledgments I am deeply indebted to countless individuals for their support and encouragement during the past five years of graduate studies. First and foremost, I would like to express my gratitude to my mentor, Dr. Eric P. Hoffman, for his unwavering support and guidance, and keen attention to my professional development. This Dissertation would not have been possible without the critical input he provided and the engaging environment he created. -

Cldn19 Clic2 Clmp Cln3
NewbornDx™ Advanced Sequencing Evaluation When time to diagnosis matters, the NewbornDx™ Advanced Sequencing Evaluation from Athena Diagnostics delivers rapid, 5- to 7-day results on a targeted 1,722-genes. A2ML1 ALAD ATM CAV1 CLDN19 CTNS DOCK7 ETFB FOXC2 GLUL HOXC13 JAK3 AAAS ALAS2 ATP1A2 CBL CLIC2 CTRC DOCK8 ETFDH FOXE1 GLYCTK HOXD13 JUP AARS2 ALDH18A1 ATP1A3 CBS CLMP CTSA DOK7 ETHE1 FOXE3 GM2A HPD KANK1 AASS ALDH1A2 ATP2B3 CC2D2A CLN3 CTSD DOLK EVC FOXF1 GMPPA HPGD K ANSL1 ABAT ALDH3A2 ATP5A1 CCDC103 CLN5 CTSK DPAGT1 EVC2 FOXG1 GMPPB HPRT1 KAT6B ABCA12 ALDH4A1 ATP5E CCDC114 CLN6 CUBN DPM1 EXOC4 FOXH1 GNA11 HPSE2 KCNA2 ABCA3 ALDH5A1 ATP6AP2 CCDC151 CLN8 CUL4B DPM2 EXOSC3 FOXI1 GNAI3 HRAS KCNB1 ABCA4 ALDH7A1 ATP6V0A2 CCDC22 CLP1 CUL7 DPM3 EXPH5 FOXL2 GNAO1 HSD17B10 KCND2 ABCB11 ALDOA ATP6V1B1 CCDC39 CLPB CXCR4 DPP6 EYA1 FOXP1 GNAS HSD17B4 KCNE1 ABCB4 ALDOB ATP7A CCDC40 CLPP CYB5R3 DPYD EZH2 FOXP2 GNE HSD3B2 KCNE2 ABCB6 ALG1 ATP8A2 CCDC65 CNNM2 CYC1 DPYS F10 FOXP3 GNMT HSD3B7 KCNH2 ABCB7 ALG11 ATP8B1 CCDC78 CNTN1 CYP11B1 DRC1 F11 FOXRED1 GNPAT HSPD1 KCNH5 ABCC2 ALG12 ATPAF2 CCDC8 CNTNAP1 CYP11B2 DSC2 F13A1 FRAS1 GNPTAB HSPG2 KCNJ10 ABCC8 ALG13 ATR CCDC88C CNTNAP2 CYP17A1 DSG1 F13B FREM1 GNPTG HUWE1 KCNJ11 ABCC9 ALG14 ATRX CCND2 COA5 CYP1B1 DSP F2 FREM2 GNS HYDIN KCNJ13 ABCD3 ALG2 AUH CCNO COG1 CYP24A1 DST F5 FRMD7 GORAB HYLS1 KCNJ2 ABCD4 ALG3 B3GALNT2 CCS COG4 CYP26C1 DSTYK F7 FTCD GP1BA IBA57 KCNJ5 ABHD5 ALG6 B3GAT3 CCT5 COG5 CYP27A1 DTNA F8 FTO GP1BB ICK KCNJ8 ACAD8 ALG8 B3GLCT CD151 COG6 CYP27B1 DUOX2 F9 FUCA1 GP6 ICOS KCNK3 ACAD9 ALG9 -

(P -Value<0.05, Fold Change≥1.4), 4 Vs. 0 Gy Irradiation
Table S1: Significant differentially expressed genes (P -Value<0.05, Fold Change≥1.4), 4 vs. 0 Gy irradiation Genbank Fold Change P -Value Gene Symbol Description Accession Q9F8M7_CARHY (Q9F8M7) DTDP-glucose 4,6-dehydratase (Fragment), partial (9%) 6.70 0.017399678 THC2699065 [THC2719287] 5.53 0.003379195 BC013657 BC013657 Homo sapiens cDNA clone IMAGE:4152983, partial cds. [BC013657] 5.10 0.024641735 THC2750781 Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (HL1). 4.07 0.04353262 DNAH5 [Source:Uniprot/SWISSPROT;Acc:Q8TE73] [ENST00000382416] 3.81 0.002855909 NM_145263 SPATA18 Homo sapiens spermatogenesis associated 18 homolog (rat) (SPATA18), mRNA [NM_145263] AA418814 zw01a02.s1 Soares_NhHMPu_S1 Homo sapiens cDNA clone IMAGE:767978 3', 3.69 0.03203913 AA418814 AA418814 mRNA sequence [AA418814] AL356953 leucine-rich repeat-containing G protein-coupled receptor 6 {Homo sapiens} (exp=0; 3.63 0.0277936 THC2705989 wgp=1; cg=0), partial (4%) [THC2752981] AA484677 ne64a07.s1 NCI_CGAP_Alv1 Homo sapiens cDNA clone IMAGE:909012, mRNA 3.63 0.027098073 AA484677 AA484677 sequence [AA484677] oe06h09.s1 NCI_CGAP_Ov2 Homo sapiens cDNA clone IMAGE:1385153, mRNA sequence 3.48 0.04468495 AA837799 AA837799 [AA837799] Homo sapiens hypothetical protein LOC340109, mRNA (cDNA clone IMAGE:5578073), partial 3.27 0.031178378 BC039509 LOC643401 cds. [BC039509] Homo sapiens Fas (TNF receptor superfamily, member 6) (FAS), transcript variant 1, mRNA 3.24 0.022156298 NM_000043 FAS [NM_000043] 3.20 0.021043295 A_32_P125056 BF803942 CM2-CI0135-021100-477-g08 CI0135 Homo sapiens cDNA, mRNA sequence 3.04 0.043389246 BF803942 BF803942 [BF803942] 3.03 0.002430239 NM_015920 RPS27L Homo sapiens ribosomal protein S27-like (RPS27L), mRNA [NM_015920] Homo sapiens tumor necrosis factor receptor superfamily, member 10c, decoy without an 2.98 0.021202829 NM_003841 TNFRSF10C intracellular domain (TNFRSF10C), mRNA [NM_003841] 2.97 0.03243901 AB002384 C6orf32 Homo sapiens mRNA for KIAA0386 gene, partial cds. -

Microtubule Organization and Microtubule- Associated Proteins (Maps)
Chapter 3 Microtubule Organization and Microtubule- Associated Proteins (MAPs) Elena Tortosa, Lukas C. Kapitein, and Casper C. Hoogenraad Abstract Dendrites have a unique microtubule organization. In vertebrates, den- dritic microtubules are organized in antiparallel bundles, oriented with their plus ends either pointing away or toward the soma. The mixed microtubule arrays control intracellular trafficking and local signaling pathways, and are essential for dendrite development and function. The organization of microtubule arrays largely depends on the combined function of different microtubule regulatory factors or generally named microtubule-associated proteins (MAPs). Classical MAPs, also called structural MAPs, were identified more than 20 years ago based on their ability to bind to and copurify with microtubules. Most classical MAPs bind along the microtubule lattice and regulate microtubule polymerization, bundling, and stabilization. Recent evidences suggest that classical MAPs also guide motor protein transport, interact with the actin cytoskeleton, and act in various neuronal signaling networks. Here, we give an overview of microtubule organization in dendrites and the role of classical MAPs in dendrite development, dendritic spine formation, and synaptic plasticity. Keywords Neuron • Dendrite • Cytoskeleton • Microtubule • Microtubule- associated protein • MAP1 • MAP2 • MAP4 • MAP6 • MAP7 • MAP9 • Tau 3.1 Introduction Microtubules (MTs) are cytoskeletal structures that play essential roles in all eukaryotic cells. MTs are important not only during cell division but also in non-dividing cells, where they are critical structures in numerous cellular processes such as cell motility, migration, differentiation, intracellular transport and organelle positioning. MTs are composed of two proteins, α- and β-tubulin, that form heterodimers and organize themselves in a head-to-tail manner. -

Mai Muudatuntuu Ti on Man Mini
MAIMUUDATUNTUU US009809854B2 TI ON MAN MINI (12 ) United States Patent ( 10 ) Patent No. : US 9 ,809 ,854 B2 Crow et al. (45 ) Date of Patent : Nov . 7 , 2017 Whitehead et al. (2005 ) Variation in tissue - specific gene expression ( 54 ) BIOMARKERS FOR DISEASE ACTIVITY among natural populations. Genome Biology, 6 :R13 . * AND CLINICAL MANIFESTATIONS Villanueva et al. ( 2011 ) Netting Neutrophils Induce Endothelial SYSTEMIC LUPUS ERYTHEMATOSUS Damage , Infiltrate Tissues, and Expose Immunostimulatory Mol ecules in Systemic Lupus Erythematosus . The Journal of Immunol @(71 ) Applicant: NEW YORK SOCIETY FOR THE ogy , 187 : 538 - 552 . * RUPTURED AND CRIPPLED Bijl et al. (2001 ) Fas expression on peripheral blood lymphocytes in MAINTAINING THE HOSPITAL , systemic lupus erythematosus ( SLE ) : relation to lymphocyte acti vation and disease activity . Lupus, 10 :866 - 872 . * New York , NY (US ) Crow et al . (2003 ) Microarray analysis of gene expression in lupus. Arthritis Research and Therapy , 5 :279 - 287 . * @(72 ) Inventors : Mary K . Crow , New York , NY (US ) ; Baechler et al . ( 2003 ) Interferon - inducible gene expression signa Mikhail Olferiev , Mount Kisco , NY ture in peripheral blood cells of patients with severe lupus . PNAS , (US ) 100 ( 5 ) : 2610 - 2615. * GeneCards database entry for IFIT3 ( obtained from < http : / /www . ( 73 ) Assignee : NEW YORK SOCIETY FOR THE genecards. org /cgi - bin / carddisp .pl ? gene = IFIT3 > on May 26 , 2016 , RUPTURED AND CRIPPLED 15 pages ) . * Navarra et al. (2011 ) Efficacy and safety of belimumab in patients MAINTAINING THE HOSPITAL with active systemic lupus erythematosus : a randomised , placebo FOR SPECIAL SURGERY , New controlled , phase 3 trial . The Lancet , 377 :721 - 731. * York , NY (US ) Abramson et al . ( 1983 ) Arthritis Rheum . -

In This Table Protein Name, Uniprot Code, Gene Name P-Value
Supplementary Table S1: In this table protein name, uniprot code, gene name p-value and Fold change (FC) for each comparison are shown, for 299 of the 301 significantly regulated proteins found in both comparisons (p-value<0.01, fold change (FC) >+/-0.37) ALS versus control and FTLD-U versus control. Two uncharacterized proteins have been excluded from this list Protein name Uniprot Gene name p value FC FTLD-U p value FC ALS FTLD-U ALS Cytochrome b-c1 complex P14927 UQCRB 1.534E-03 -1.591E+00 6.005E-04 -1.639E+00 subunit 7 NADH dehydrogenase O95182 NDUFA7 4.127E-04 -9.471E-01 3.467E-05 -1.643E+00 [ubiquinone] 1 alpha subcomplex subunit 7 NADH dehydrogenase O43678 NDUFA2 3.230E-04 -9.145E-01 2.113E-04 -1.450E+00 [ubiquinone] 1 alpha subcomplex subunit 2 NADH dehydrogenase O43920 NDUFS5 1.769E-04 -8.829E-01 3.235E-05 -1.007E+00 [ubiquinone] iron-sulfur protein 5 ARF GTPase-activating A0A0C4DGN6 GIT1 1.306E-03 -8.810E-01 1.115E-03 -7.228E-01 protein GIT1 Methylglutaconyl-CoA Q13825 AUH 6.097E-04 -7.666E-01 5.619E-06 -1.178E+00 hydratase, mitochondrial ADP/ATP translocase 1 P12235 SLC25A4 6.068E-03 -6.095E-01 3.595E-04 -1.011E+00 MIC J3QTA6 CHCHD6 1.090E-04 -5.913E-01 2.124E-03 -5.948E-01 MIC J3QTA6 CHCHD6 1.090E-04 -5.913E-01 2.124E-03 -5.948E-01 Protein kinase C and casein Q9BY11 PACSIN1 3.837E-03 -5.863E-01 3.680E-06 -1.824E+00 kinase substrate in neurons protein 1 Tubulin polymerization- O94811 TPPP 6.466E-03 -5.755E-01 6.943E-06 -1.169E+00 promoting protein MIC C9JRZ6 CHCHD3 2.912E-02 -6.187E-01 2.195E-03 -9.781E-01 Mitochondrial 2- -

Pubertal Mouse Mammary Gland Development - Transcriptome Analysis and the Investigation of Fbln2 Expression and Function
Olijnyk, Daria (2011) Pubertal mouse mammary gland development - transcriptome analysis and the investigation of Fbln2 expression and function. PhD thesis. http://theses.gla.ac.uk/2996/ Copyright and moral rights for this thesis are retained by the author A copy can be downloaded for personal non-commercial research or study, without prior permission or charge This thesis cannot be reproduced or quoted extensively from without first obtaining permission in writing from the Author The content must not be changed in any way or sold commercially in any format or medium without the formal permission of the Author When referring to this work, full bibliographic details including the author, title, awarding institution and date of the thesis must be given Glasgow Theses Service http://theses.gla.ac.uk/ [email protected] Pubertal Mouse Mammary Gland Development- Transcriptome Analysis and the Investigation of Fbln2 Expression and Function Daria Olijnyk (B.Sc, MRes) Thesis submitted to the University of Glasgow in fulfilment of the requirements for the degree of Doctor of Philosophy June 2011 Institute of Cancer Sciences College of Medical, Veterinary and Life Sciences University of Glasgow 2 For my parents, Bogusława and Bogusław Olijnyk and grandmother, Anna Bandurak 3 Abstract Mouse mammary gland morphogenesis at puberty is a complex developmental process, regulated by systemic hormones, local growth factors and dependent on the epithelial/epithelial and epithelial/stromal interactions. TEBs which invade the fat pad are important in laying down the epithelial framework of the gland at this time point. The objective of this thesis was to use a combination of „pathway-„ and „candidate gene analysis‟ of the transcriptome of isolated TEBs and ducts and associated stroma, combined with detailed analyses of selected proteins, to further define the key proteins and processes involved at puberty. -

Role of the Microtubule-Associated Protein ATIP3 in Cell Migration and Breast Cancer Metastasis Angie Molina Delgado
Role of the microtubule-associated protein ATIP3 in cell migration and breast cancer metastasis Angie Molina Delgado To cite this version: Angie Molina Delgado. Role of the microtubule-associated protein ATIP3 in cell migration and breast cancer metastasis. Molecular biology. Université René Descartes - Paris V, 2014. English. NNT : 2014PA05T022. tel-01068663 HAL Id: tel-01068663 https://tel.archives-ouvertes.fr/tel-01068663 Submitted on 26 Sep 2014 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Université Paris Descartes Ecole doctorale BioSPC Thesis submitted towards fulfillment of the requirement for the degree of DOCTOR of Health & Life Sciences Specialized in Cellular and Molecular Biology Role of the microtubule-associated protein ATIP3 in cell migration and breast cancer metastasis By Angie Molina Delgado Under supervision of Dr. Clara Nahmias Thesis defense 3 September, 2014 Members of jury: Dr. Ali BADACHE Reviewer Dr. Laurence LAFANECHERE Reviewer Dr. Franck PEREZ Examiner Dr. Stéphane HONORE Examiner Dr. Clara NAHMIAS Thesis Director Table of Contents List of abbreviations ....................................................................................................................... 9 “Rôle de la protéine associée aux microtubules ATIP3 dans la migration cellulaire et la formation de métastases du cancer du sein” ................................................................................................ 11 I. -

A Distinct Inflammatory Gene Expression Profile in Patients with Psoriatic Arthritis
Genes and Immunity (2006) 7, 583–591 & 2006 Nature Publishing Group All rights reserved 1466-4879/06 $30.00 www.nature.com/gene ORIGINAL ARTICLE A distinct inflammatory gene expression profile in patients with psoriatic arthritis AK Stoeckman1, EC Baechler1, WA Ortmann1, TW Behrens1, CJ Michet2 and EJ Peterson1 1Department of Medicine, University of Minnesota Medical School, Center for Immunology, University of Minnesota, Minneapolis, MN, USA and 2Division of Rheumatology, Mayo Clinic, Rochester, MN, USA Psoriatic arthritis (PsA) is a systemic inflammatory condition featuring polyarthritis associated with psoriasis. Apart from clinical indicators, few biomarkers exist to aid in the diagnosis and management of PsA. We hypothesized that whole blood gene expression profiling would provide new diagnostic markers and/or insights into pathogenesis of the disease. We compared whole blood gene expression profiles in PsA patients and in age-matched controls. We identified 310 differentially expressed genes, the majority of which are upregulated in PsA patients. The PsA expression profile does not significantly overlap with profiles derived from patients with rheumatoid arthritis or systemic lupus erythematosus. Logistic regression identified two lymphocyte-specific genes (zinc-finger protein 395 and phosphoinositide-3-kinase 2B) that discriminate PsA patients from normal controls. In addition, a highly coregulated cluster of overexpressed genes implicated in protein kinase A regulation strongly correlates with erythrocyte sedimentation rate. Other clusters of coregulated, yet suppressed genes in PsA patient blood include molecules involved in T-cell signaling. Finally, differentially expressed genes in PsA fall into diverse functional categories, but many downregulated genes belong to a CD40 signaling pathway. Together, the data suggest that gene expression profiles of PsA patient blood contain candidate novel disease markers and clues to pathogenesis.