Identification Genes Controlling Capsaicinoid Content in Capsicum
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Epicurean Product Listing 2016
Epicurean Product Listing 2016 800.934.6495 173 Thorn Hill Rd Warrendale, PA 15086 ** For the most up to date listing, please visit our website ** version 2, 8/26/16 EPICUREAN PRODUCT LISTING Condiments.........................................3 Miscellaneous......................................9 Oils & Vinegars................................1211 Syrups.............................................1514 Spices.............................................1716 Dried Mushrooms............................2322 Dried Fruits & Nuts..........................2423 Breads and Crackers.......................2726 Meats & Seafood.............................3029 Pasta Sauces and Noodles.............3332 Desserts..........................................3635 Chocolate........................................4039 Grains & Legumes...........................4342 Cheese, Dairy, & Eggs....................4544 Bar & Bakery.......................................50 Baking & Pastry...................................54 Appetizers...........................................66 CONDIMENTS Prod # Description Packaging UoM Special Order 06177 BASE BEEF NO MSG 1 LB EA X 06207 BASE BEEF NO MSG 5 LB EA 06176 BASE BEEF NO MSG MINORS 12/1 LB CS X 06206 BASE BEEF NO MSG MINORS 4/5 LB CS 06179 BASE CHICKEN NO MSG 1 LB EA 06201 BASE CHICKEN NO MSG 5 LB EA 06178 BASE CHICKEN NO MSG MINORS 12/1 LB CS 06200 BASE CHICKEN NO MSG MINORS 4/5 LB CS 06180 BASE CLAM NO MSG MINORS 6/1 LB CS 06198 BASE CRAB NO MSG MINORS 6/1 LB CS 06187 BASE ESPAGNOLE SAUCE 1 LB EA 06186 BASE ESPAGNOLE SAUCE -

A Determination of the Physical, Chemical, and Biological Features Of
Louisiana State University LSU Digital Commons LSU Master's Theses Graduate School 2008 A determination of the physical, chemical, and biological features of suspended dark flecks in hot sauce Andre Brock Louisiana State University and Agricultural and Mechanical College Follow this and additional works at: https://digitalcommons.lsu.edu/gradschool_theses Recommended Citation Brock, Andre, "A determination of the physical, chemical, and biological features of suspended dark flecks in hot sauce" (2008). LSU Master's Theses. 1035. https://digitalcommons.lsu.edu/gradschool_theses/1035 This Thesis is brought to you for free and open access by the Graduate School at LSU Digital Commons. It has been accepted for inclusion in LSU Master's Theses by an authorized graduate school editor of LSU Digital Commons. For more information, please contact [email protected]. A DETERMINATION OF THE PHYSICAL, CHEMICAL, AND BIOLOGICAL FEATURES OF SUSPENDED DARK FLECKS IN HOT SAUCE A Thesis Submitted to the Graduate Faculty of the Louisiana State University and Agricultural and Mechanical College in partial fulfillment of the requirements for the degree of Master of Science in Horticulture by André Brock B.S., Louisiana State University, 2000 December 2008 ACKNOWLEDGEMENTS I owe a debt of gratitude to Dr. Paul Wilson, whose support made this project possible. His expertise and patience will always be appreciated. Thanks also to the rest of my committee. Dr. Carl Motsenbocker, Dr. Marlene Janes, and Dr. Jack Losso were all instrumental in the completion of the project. Their guidance in content and in scientific techniques in various fields was important in forming a well-rounded education. -

Name Description Type Culinary Uses Flavor Scoville Matures
Name Description Type Culinary Uses Flavor Scoville Matures Baby Chocolate Bell Gourmet miniature that are 2 1/2" tall and 1 Bell Stuffed, pickled, Sweet 0 - 100 85 1/2" wide, with all the flavor of full-sized bells. canned, salads or They will mature from green to the color of milk fresh eating chocolate. The mature peppers are the sweetest. These compact plants are amazingly productive. Beaver Dam Yields enormous amounts of horn shaped, Bell Fresh eating, roasting, Mild, Sweet 500-1,000 80 medium-hot peppers on compact plants. Great salsa, pickled or stuffing pepper that ripens from green to red. stuffed This pepper will have more heat when seeded, when cooked the heat mellows but it will retain its robust flavor. Better Belle IV Crisp, blocky, thick walled shiny fruit that are full Bell Great for stuffing, Sweet 0-100 75 of flavor. They ripen from green to shiny red. roasting, grilled, This one has a better production than the canning , drying or original. freezing. Big Bertha Produces thick, crisp peppers that are 7" long Bell Excellent for stuffing, Sweet, crisp 0 - 100 72 and extremely sweet, with few seeds. They roasting, salads or mature from dark green to shiny red. For best snacks flavor, eat them the same day that they are picked. Cajun Bell Produces small 2-3" long lobed peppers with the Bell Can be stuffed, also Spicy, hint of 100 - 1,000 60 flavor of a sweet pepper along with a mild, spicy adds color and flavor sweet heat. They ripen from green to orange to red. -

111 Ideas for Innovation Surprising Sides Beers
NO. 48 FULL-SERVICE RESTAURANTS : SETTING AMERICA’S TABLE ® Beers for 2018 111 Ideas for Innovation Cold-Brew Cocktails Surprising Sides Urban Cowboy Kimbal Musk sits down with FSR and explains his strategy to disrupt the casual-dining segment with urban casual. BEST PRACTICES 111 Ideas for Menu Innovation +PITGFKGPVUƃCXQTUHQQFUDGXGTCIGUtCNN VJGDGUVKFGCUHTQOEJGHUCPFTGUVCWTCPVU CTQWPFVJGEQWPVT[ BY AMANDA BALTAZAR, CONNIE GENTRY, AMELIA LEVIN, BEVERLY STEPHEN THINKSTOCK 50 NOVEMBER 2017 FOODNEWSFEED.COM BEST PRACTICES ACCESSORIZE WITH SAUCES AND SIDES: 1. SAVORY YOGURT 5. AÇAI Carl Jorgensen, director of Touted as a food superhero, thought leadership with açai is appearing every- Daymon, believes we’ll see where. The most popular savory yogurt take off in preparation is açai bowls— sandwich sauces and sides. smoothies containing ber- ries, nuts, grains, oatmeal, 2. ROMESCO SAUCE and their ilk. An orange sauce made with roasted red peppers, 6. SAMBAL romesco is gaining traction. Especially popular in Indo- Traditionally served with nesia and Malaysia, sam- fish, at Gato in New York bal is a sauce with a kick. City it’s served with eggs Its ingredients vary, and so EMMER & RYE and with a pork chop. do its uses. Dish it up with noodles, eggs, or as a dip. 10. FRESHLY MILLED GRAINS 3. BARBERRIES (KTUVVJG[OCFGVJGRCUVCKPJQWUGVJGPVJG[DCMGF Tagged by McCormick as a 7. GOCHUJANG their own bread; now, more chefs are milling their own flavor-leader, tart barberries This fermented Korean chili ƃQWT%JGH-GXKP(KPMCV'OOGT4[GKP#WUVKP6GZCU can be used in sweet and paste can be used to add is a leading proponent. The restaurant is named after savory dishes, and are rec- heat and flavor. -

Hot Pepper (Capsicum Spp.) – Important Crop on Guam
Food Plant Production June 2017 FPP-05 Hot Pepper (Capsicum spp.) – Important Crop on Guam Joe Tuquero, R. Gerard Chargualaf and Mari Marutani, Cooperative Extension & Outreach College of Natural & Applied Sciences, University of Guam Most Capsicum peppers are known for their spicy heat. Some varieties have little to no spice such as paprika, banana peppers, and bell peppers. The spice heat of Capsicum peppers are measured and reported as Scoville Heat Units (SHU). In 1912, American pharmacist, Wilbur Scoville, developed a test known as the, Scoville Organoleptic Test, which was used to measure pungency (spice heat) of Capsicum peppers. Since the 1980s, pungency has been more accurately measured by high-performance liquid chromatography Source: https://phys.org/news/2009-06-domestication- (HPLC). HPLC tests result in American Spice Trade capsicum-annuum-chile-pepper.html Association (ASTA) pungency units. ASTA pungency Introduction units can be converted to SHU. Table 2 displays Sco- Hot pepper, also known as chili, chilli, or chile pepper, ville Heat Units of various popular Capsicum peppers is a widely cultivated vegetable crop that originates (Wikipedia, 2017). from Central and South America. Hot peppers belong to the genus Capsicum. There are over 20 species under the genus Capsicum. There are five major domesticated species of peppers that are commercially cultivated (Table 1), and there are more than 50,000 varieties. Fig. 1 depicts a unqiue, citrus-flavored variety of Capsicum baccatum hot pepper, known as Lemon Drop (aji-type), popular for seasoning in Peru (Wikipedia, 2017). Table 1. The five major domesticated Capsicum species of pepper with examples of commonly known types of pepper. -

Supercritical Carbon Dioxide Extraction of Oleoresin from Malagueta Pepper (Capsicum Frutescens L.) Enhanced by Ultrasound
III Iberoamerican Conference on Supercritical Fluids Cartagena de Indias (Colombia), 2013 SUPERCRITICAL CARBON DIOXIDE EXTRACTION OF OLEORESIN FROM MALAGUETA PEPPER (CAPSICUM FRUTESCENS L.) ENHANCED BY ULTRASOUND Philipe dos Santos¹, Ana C. de Aguiar¹, Camila A. Rezende2 and Julian Martínez¹* ¹Food Engineering Departament, Food Engineering College University of Campinas (UNICAMP) R. Monteiro Lobato 80, P.O. Box:6121, 13083-862, Campinas, SP, Brazil 2Institute of Chemistry University of Campinas (UNICAMP) Campinas, SP, Brazil Email: [email protected] Abstract. Supercritical fluid extraction technology came as an alternative to traditional methods of extraction and fractionation of bioactive compounds. The capacity of a supercritical fluid extraction unit is usually changed through the application of combined techniques, such as the use of different co-solvents and ultrasonic waves. The ultrasonic technology is based on the high frequency ultrasonic waves, which are capable of causing cavitations and disrupting the cell walls of vegetable materials. This favors the penetration of solvent and mass transfer, increasing the extraction yield and velocity. The objective of this study was to obtain extracts of malagueta pepper (Capsicum frutescens L.) using supercritical fluid extraction (SFE) assisted by ultrasound. The raw material used was malagueta pepper dried at 50°C (5% w. b.) and triturated. The particles with mean diameters between 1.68 to 1.18 mm and 0.342 to 0.177 mm were selected to SFE. To study the influence of ultrasonic waves and particle diameter in the extraction rate and yield, extractions were performed without ultrasound, and with ultrasound at power of 360 W. The extraction unit was constructed with the ultrasonic transducer installed inside the supercritical fluid extractor, which works in a frequency of 20 kHz and a maximum power of 800 W. -

Fresh Chiles SWEET VEGETABLES
Jalapeño, meets Fresh Three Pea, and Shrimp chiles Stir-Fry SWEET VEGETABLES Many fresh chiles When H are so hot that their flavor gets blunted by the burn. But vegetables that are high in natural sugars have an amazing taming effect. Pair the two and sud- denly chiles become brighter, less fiery— you can actually taste the layers of SWEET flavors. Peas, sweet bell peppers, and zucchini are excel- lent springtime and summertime matches, and butter- nut squash and root veggies like beets, rutabagas, and pars- nips work perfectly in the fall and winter. DELICIOUS THINGS A HAPPEN, WHICH IS WHY THIS DYNAMIC DUO HAS ROOTS IN CUISINES AROUND THE WORLD. HEAT INTENSIFIES THE FLAVORS OF SWEET INGREDIENTS, WHILE SUGARS TAME SPICY SIMMERED GRILLED FOODS AND HIGHLIGHT Blau Bette styling by Jamie Kimm; prop by styling Food CUT THEM IN HALF T SET THEM OVER RAW AND STIR INTO FRIED THEIR FRUITY NOTES. THE HOT COALS 4 ways CHOP IN SALSAS, BRAISES ALONG SLICE INTO WHOLE, THEN SLICE HERE, PERFECT PAIRINGS SHAVE INTO SALADS, WITH ONIONS OR STIR-FRIES OR AND TOSS WITH AND THE INFINITE WAYS to eat BLEND INTO SUBMERGE WHOLE A QUICK VEGETABLE OTHER GRILLED VINAIGRETTES. ONES INTO STEWS SAUTÉ. YOU CAN PLAY, FOR chiles VEGGIES OR PUT OR CHILIS (REMOVE DISHES YOU’LL WANT TO THEM ON BURGERS. CREATE ALL SUMMER... BEFORE SERVING). MAKE THAT ALL YEAR. By Genevieve Ko Photographs by Beth Galton PREP TIP To make fresh chiles milder, remove the seeds and slice out the ribs. Be sure to wear gloves—they’ll keep the peppers’ hot compounds from burning your skin. -

Pepper Joes Seeds 2017.Pdf
Maynard, MA 01754 300. Suite St., Parker 141 16 NEW HOT NEW HOT 16 THIS YEAR THIS Reapers Festival PEPPERS Wow! Not just the legendary Carolina Reaper, but now we have more in the family! If you are a Reaper fan, get ready! Carolina Reaper Grow the legendary Guinness Book of World Records hottest pepper on the planet. This is the REAL deal, from the original strain of award-winning peppers. 1,569,000 Scoville Heat Units. $9.99 (10+ seeds) Chocolate Reaper Mmmm… smoky! This delicious hot pepper tastes as good as the classic, but with the hint of a smoky taste up front. It is still being bred out for stabil- ity, but worth the taste! $9.99 (10+ seeds) of 7342companies reviewed and 30 “Top a out Company” Joe’s #1 in Pepper Seeds Dave’s Garden Ranks Pepper Yellow Reaper Try this beauty with grilled seafood! It has a fruity flavor paired with loads of heat. We are still growing this pepper out, but wanted to bring it to you without delay! US POSTAGE Sudbury, MA Sudbury, Permit No 3 $9.99 (10+ seeds) STD PRSRT PAID About Pepper Joe’s Butch “T” Trinidad ScorpionOUR W e a#3re the expWe’reerts in thrilledHot Pe topp haveer Se thiseds .# 3 “WORLD’S HOTTEST PEPPER. It set a Guinness Book PLEDGEof World Record N THS EAR - 1 SCREAMING Since 1989, Pepperearly Joe’s in 2011has found, at 1,450,000 grown, Scoville Units. WOW! NEHOT oetos EPPS and enjoyed superThat’s hot peppersa lot of fromheat. all This over is a very exclusive pepper the world. -

Genus Species/Common Names Report Genus/Species Common Name
Genus Species/Common Names Report Genus/Species Common Name Abeliophyllum Distichum White-forsythia Abelmoschus Esculentus Okra Abelmoschus Manihot Manioc-hibiscus Sunset-hibiscus Abies Alba European Silver Fir Silver Fir White Fir Abies Balsamea American Silver Fir Balm of Gilead Balsam Canada Balsam Fir Eastern Fir Abies Concolor Colorado Fir Colorado White Fir Silver Fir White Fir Abies Grandis Giant Fir Grand Fir Lowland Fir Lowland White Fir Silver Fir White Fir Yellow Fir Abies Homolepis Nikko Fir Abies Koreana Korean Fir Abies Pectinata Silver Fir Abies Sachalinensis Sakhalin Fir Abies Sibirica Siberian Fir Abies Veitchii Christmastree Veitch Fir Thursday, January 12, 2017 Page 1 of 229 Genus Species/Common Names Report Genus/Species Common Name Abies Veitchii Veitch's Silver Fir Abronia Villosa Desert Sand-verbena Abrus Fruticulosus No common names identified Abrus Precatorius Coral-beadplant Crab's-eye Indian-licorice Jequirity Jequirity-bean Licorice-vine Love-bean Lucky-bean Minnie-minnies Prayer-beads Precatory Precatory-bean Red-beadvine Rosary-pea Weatherplant Weathervine Acacia Arabica Babul Acacia Egyptian Acacia Indian Gum-arabic-tree Scented-thorn Thorn-mimosa Thorny Acacia Acacia Catechu Black Cutch Catechu Acacia Concinna Soap-pod Acacia Dealbata Mimosa Silver Wattle Acacia Decurrens Green Wattle Acacia Farnesiana Cassie Huisache Thursday, January 12, 2017 Page 2 of 229 Genus Species/Common Names Report Genus/Species Common Name Acacia Farnesiana Opopanax Popinac Sweet Acacia Acacia Mearnsii Black Wattle Tan Wattle -

Numex Lotalutein, a Lutein-Rich Serrano Pepper
HORTSCIENCE 55(12):2052–2055. 2020. https://doi.org/10.21273/HORTSCI14949-20 segregating population produced segregating lines of mature fruit that were red, yellow, and orange. Single-plant selection using phe- NuMex LotaLutein, a Lutein-rich notypic recurrent selection with pedigree breeding for earliness, desirable fruit shape Serrano Pepper and size, yield, and ease of destemming was accomplished. A total of seven generations of Ivette Guzman, Danise Coon, Krystal Vargas, and Paul W. Bosland self-pollination were accomplished using a Department of Plant and Environmental Sciences, New Mexico State greenhouse during the winter season. Within University, Las Cruces, NM 88003 the segregating generations, five single-plant selections were made based on the color of Additional index words. Capsicum annuum , carotenoids, high-performance liquid chroma- ripeness alone. Each selected plant was selfed tography, human health, macular degeneration, open-pollinated by stripping fruits and open flowers and then placing an isolation cage over the individual plant to exclude any outcrossing (Bosland, Lutein is a carotenoid with antioxidant modification by the human body (Johnson, 1993). During each generation of selection, and anti-inflammatory properties important 2002). Sources of lutein are green leafy phenotypic traits considered to be impor- for reducing the risks of several chronic vegetables, yellow fruits and vegetables, tant to a serrano pod-type were selected diseases (Buscemi et al., 2018). Biofortifica- and egg yolks (Johnson, 2002). Furthermore, (Bosland and Votava, 2012). In 2013 and tion is the process by which the nutritional many studies have reported that lutein may 2014, selfed, single-plant selections were quality of food crops, like lutein, is improved have positive effects on different clinical planted at the LPSRC, and selections of through plant breeding, genetic medication, conditions, thus ameliorating cognitive func- desired horticultural traits were made and and other agronomic practices. -
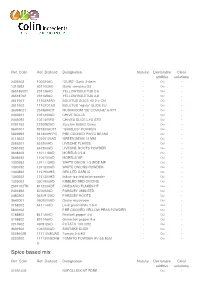
Download the Result
Ref. Colin Ref. Diafood Designation Natural Declarable Clear additive solutions 2408502 100024AD "OURS" Garlic 2-4mm - Oui - 1310592 5311G3AD Garlic semolina G3 - Oui - 36518502T 231126AD YELLOW BOLETUS 2-6 - Oui - 36518702 231169AD YELLOW BOLETUS 3-8 - Oui - 3611601 11102A2AD BOLETUS SLICE A2 2-4 CM - Oui - 3611602 11102O1AD BOLETUS "edulis" SLIDE EU - Oui - 3638902T 251969ADT MUSHROOM "DE COUCHE" 6-9TT - Oui - 2443201 1021400AD CHIVE ROLLS - Oui - 2445092 1021400FD CHIVES SLICE LYO STD - Oui - 0091162 1230982AD Zucchini 8X8X2 China - Oui - 3640101 921300ADTT "GIROLES" POWDER - Oui - 5823992 941300HFPC PRE COOKED PINTO BEANS - Oui - 0113602 10100125AD GREEN BEAN 12 MM - Oui - 2585201 651200AD LIVECHE FLAKES - Oui - 2585192 651300AD LIVECHE ROOTS POWDER - Oui - 3648403 1101118AD MORELS 0.5-8 - Oui - 3648492 1100700AD MORELS MP - Oui - 1330592 1241113AD WHITE ONION 1-3 INDE MP - Oui - 1330192 1241300AD WHITE ONIONS POWDER - Oui - 1340892 1151900HS GRILLED GARLIC - Oui - 1340202 1151300HS Indian toasted onion powder - Oui - 1326002 3501900AD KIBBLED RED ONIONS - Oui - 2591702TK 811200ADT OREGANO FLAKES HT - Oui - 2491892 870022AD PARSLEY 2MM STD - Oui - 2480902 3670912AD PARSLEY ROOTS - Oui - 3642001 0502000AD Oyster mushroom - Oui - 0158202 641114AD Leek green/white 1.5-4 - Oui - 5830102 PRE COOKED YELLOW PEAS POWDER - Oui - 0188802 841146AD Red bell pepper 4-6 - Oui - 0198802 831146AD Green bell pepper 4-6 - Oui - 0210902 490912AD POTATO 10X10X2 - Oui - 3692902 1040200AD SHIITAKE SLICE - Oui - 0228402E 1171124EUAD Tomato 2-4 EU - Oui - 0220202 1171300SDHB TOMATO POWDER HV SS E551 - Oui - O Spice based mix Ref. Colin Ref. Diafood Designation Natural Declarable Clear additive solutions 010A1402 NAPOLI MIX NF PDRE - Oui - Flavourings & Food Colourings Ref. Colin Ref. Diafood Designation Natural Declarable Clear additive solutions 284R17051 B SEASONING CURRY - - - 301U2001 LIME FLAVOUR NA Oui - - 600012102 Basil and tomato Edamame mix - Oui - Flavours Ref. -

Updates and Observations on High Tunnel Production at DSAC
2015 HIGH TUNNEL HEIRLOOM TOMATO & SPECIALTY HOT PEPPER VARIETY TRIALS Shelby Henning Department of Crop Sciences University of Illinois UNIVERSITY OF ILLINOIS RESEARCH AND EDUCATION CENTERS * UNIVERSITY OF ILLINOIS RESEARCH AND EDUCATION * CENTERS * XXXXX XXXXX XXXXXX UNIVERSITY OF ILLINOIS RESEARCH X AND EDUCATION CENTERS XXXXXXXX XXXXXXXXX 2015 HT HEIRLOOM TOMATO TRIAL • Heirloom tomato • Advantages • Flavor • Unique appearance • Profitability • Disadvantages • Tender • Performance issues 2015 HT HEIRLOOM TOMATO TRIAL • Heirloom tomato • Health benefits? • Lycopene - anticancer • Improved transportation & appearance?? • Breaker vs. vine ripe? • HT performance in N IL? www.tasti-lee.com http://www.burpee.com/vegetables/tomatoes/saladette/tomato- health-kick-hybrid-prod000992.html 2015 HT HEIRLOOM TOMATO TRIAL • 22 tomato cultivars Lycopene vs. polyphenols • 20 indeterminate • 2 determinate • 8 Replicates • - 4 analyzed as breakers • - 4 analyzed as vine-ripe • Seeded March 18 • Planted May 9 • Weekly harvests (7/30 – 9/9) 2015 HT TOMATO CULTIVAR TRIAL • Indeterminate • Spaced at 18” in row spacing • Pruned to a single leader • Trained onto single twine • Determinate • Spaced at 18 inches • Pruned to first sucker below first cluster • Trained onto single twine • Fertigated weekly with calcium nitrate and potassium nitrate @ 7 lb 8 oz. N/A TOMATO VARIETIES • Amish Gold Slicer • Gold Medal • Pareso • Arkansas Traveler • Green Zebra • Ponderosa Red • Aunt Ruby's German Green • Health Kick • Sakura • Big beef • Hillbilly Potato Leaf • Stupice • Black Krim • Japanese Black Trifele • Tasti-Lee • Brandywine • Kellog's Breakfast • Cherokee Purple • Mortgage Lifter • Favorita • Nyagous • German Pink 2015 HT TOMATO & BELL PEPPER CULTIVAR TRIAL • Insect Pest Problems • Hornworm • Mosquitos • Disease Problems • No biotic disease that appeared significant July 23, 2015 Sept.