Leptospira Proteins Identified in Leptospira In
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

METACYC ID Description A0AR23 GO:0004842 (Ubiquitin-Protein Ligase
Electronic Supplementary Material (ESI) for Integrative Biology This journal is © The Royal Society of Chemistry 2012 Heat Stress Responsive Zostera marina Genes, Southern Population (α=0. -

A Putative Cystathionine Beta-Synthase Homolog of Mycolicibacterium Smegmatis Is Involved in De Novo Cysteine Biosynthesis
University of Arkansas, Fayetteville ScholarWorks@UARK Theses and Dissertations 5-2020 A Putative Cystathionine Beta-Synthase Homolog of Mycolicibacterium smegmatis is Involved in de novo Cysteine Biosynthesis Saroj Kumar Mahato University of Arkansas, Fayetteville Follow this and additional works at: https://scholarworks.uark.edu/etd Part of the Cell Biology Commons, Molecular Biology Commons, and the Pathogenic Microbiology Commons Citation Mahato, S. K. (2020). A Putative Cystathionine Beta-Synthase Homolog of Mycolicibacterium smegmatis is Involved in de novo Cysteine Biosynthesis. Theses and Dissertations Retrieved from https://scholarworks.uark.edu/etd/3639 This Thesis is brought to you for free and open access by ScholarWorks@UARK. It has been accepted for inclusion in Theses and Dissertations by an authorized administrator of ScholarWorks@UARK. For more information, please contact [email protected]. A Putative Cystathionine Beta-Synthase Homolog of Mycolicibacterium smegmatis is Involved in de novo Cysteine Biosynthesis A thesis submitted in partial fulfillment of the requirement for the degree of Master of Science in Cell and Molecular Biology by Saroj Kumar Mahato Purbanchal University Bachelor of Science in Biotechnology, 2016 May 2020 University of Arkansas This thesis is approved for recommendation to the Graduate Council. ___________________________________ Young Min Kwon, Ph.D. Thesis Director ___________________________________ ___________________________________ Suresh Thallapuranam, Ph.D. Inés Pinto, Ph.D. Committee Member Committee Member ABSTRACT Mycobacteria include serious pathogens of humans and animals. Mycolicibacterium smegmatis is a non-pathogenic model that is widely used to study core mycobacterial metabolism. This thesis explores mycobacterial pathways of cysteine biosynthesis by generating and study of genetic mutants of M. smegmatis. Published in vitro biochemical studies had revealed three independent routes to cysteine synthesis in mycobacteria involving separate homologs of cysteine synthase, namely CysK1, CysK2, and CysM. -

Association Between the Gut Microbiota and Blood Pressure in a Population Cohort of 6953 Individuals
Journal of the American Heart Association ORIGINAL RESEARCH Association Between the Gut Microbiota and Blood Pressure in a Population Cohort of 6953 Individuals Joonatan Palmu , MD; Aaro Salosensaari , MSc; Aki S. Havulinna , DSc (Tech); Susan Cheng , MD, MPH; Michael Inouye, PhD; Mohit Jain, MD, PhD; Rodolfo A. Salido , BSc; Karenina Sanders , BSc; Caitriona Brennan, BSc; Gregory C. Humphrey, BSc; Jon G. Sanders , PhD; Erkki Vartiainen , MD, PhD; Tiina Laatikainen , MD, PhD; Pekka Jousilahti, MD, PhD; Veikko Salomaa , MD, PhD; Rob Knight , PhD; Leo Lahti , DSc (Tech); Teemu J. Niiranen , MD, PhD BACKGROUND: Several small-scale animal studies have suggested that gut microbiota and blood pressure (BP) are linked. However, results from human studies remain scarce and conflicting. We wanted to elucidate the multivariable-adjusted as- sociation between gut metagenome and BP in a large, representative, well-phenotyped population sample. We performed a focused analysis to examine the previously reported inverse associations between sodium intake and Lactobacillus abun- dance and between Lactobacillus abundance and BP. METHODS AND RESULTS: We studied a population sample of 6953 Finns aged 25 to 74 years (mean age, 49.2±12.9 years; 54.9% women). The participants underwent a health examination, which included BP measurement, stool collection, and 24-hour urine sampling (N=829). Gut microbiota was analyzed using shallow shotgun metagenome sequencing. In age- and sex-adjusted models, the α (within-sample) and β (between-sample) diversities of taxonomic composition were strongly re- lated to BP indexes (P<0.001 for most). In multivariable-adjusted models, β diversity was only associated with diastolic BP (P=0.032). -

Yeast Genome Gazetteer P35-65
gazetteer Metabolism 35 tRNA modification mitochondrial transport amino-acid metabolism other tRNA-transcription activities vesicular transport (Golgi network, etc.) nitrogen and sulphur metabolism mRNA synthesis peroxisomal transport nucleotide metabolism mRNA processing (splicing) vacuolar transport phosphate metabolism mRNA processing (5’-end, 3’-end processing extracellular transport carbohydrate metabolism and mRNA degradation) cellular import lipid, fatty-acid and sterol metabolism other mRNA-transcription activities other intracellular-transport activities biosynthesis of vitamins, cofactors and RNA transport prosthetic groups other transcription activities Cellular organization and biogenesis 54 ionic homeostasis organization and biogenesis of cell wall and Protein synthesis 48 plasma membrane Energy 40 ribosomal proteins organization and biogenesis of glycolysis translation (initiation,elongation and cytoskeleton gluconeogenesis termination) organization and biogenesis of endoplasmic pentose-phosphate pathway translational control reticulum and Golgi tricarboxylic-acid pathway tRNA synthetases organization and biogenesis of chromosome respiration other protein-synthesis activities structure fermentation mitochondrial organization and biogenesis metabolism of energy reserves (glycogen Protein destination 49 peroxisomal organization and biogenesis and trehalose) protein folding and stabilization endosomal organization and biogenesis other energy-generation activities protein targeting, sorting and translocation vacuolar and lysosomal -

Proteome Cold-Shock Response in the Extremely Acidophilic Archaeon, Cuniculiplasma Divulgatum
microorganisms Article Proteome Cold-Shock Response in the Extremely Acidophilic Archaeon, Cuniculiplasma divulgatum Rafael Bargiela 1 , Karin Lanthaler 1,2, Colin M. Potter 1,2 , Manuel Ferrer 3 , Alexander F. Yakunin 1,2, Bela Paizs 1,2, Peter N. Golyshin 1,2 and Olga V. Golyshina 1,2,* 1 School of Natural Sciences, Bangor University, Deiniol Rd, Bangor LL57 2UW, UK; [email protected] (R.B.); [email protected] (K.L.); [email protected] (C.M.P.); [email protected] (A.F.Y.); [email protected] (B.P.); [email protected] (P.N.G.) 2 Centre for Environmental Biotechnology, Bangor University, Deiniol Rd, Bangor LL57 2UW, UK 3 Systems Biotechnology Group, Department of Applied Biocatalysis, CSIC—Institute of Catalysis, Marie Curie 2, 28049 Madrid, Spain; [email protected] * Correspondence: [email protected]; Tel.: +44-1248-388607; Fax: +44-1248-382569 Received: 27 April 2020; Accepted: 15 May 2020; Published: 19 May 2020 Abstract: The archaeon Cuniculiplasma divulgatum is ubiquitous in acidic environments with low-to-moderate temperatures. However, molecular mechanisms underlying its ability to thrive at lower temperatures remain unexplored. Using mass spectrometry (MS)-based proteomics, we analysed the effect of short-term (3 h) exposure to cold. The C. divulgatum genome encodes 2016 protein-coding genes, from which 819 proteins were identified in the cells grown under optimal conditions. In line with the peptidolytic lifestyle of C. divulgatum, its intracellular proteome revealed the abundance of proteases, ABC transporters and cytochrome C oxidase. From 747 quantifiable polypeptides, the levels of 582 proteins showed no change after the cold shock, whereas 104 proteins were upregulated suggesting that they might be contributing to cold adaptation. -

Supplemental Methods
Supplemental Methods: Sample Collection Duplicate surface samples were collected from the Amazon River plume aboard the R/V Knorr in June 2010 (4 52.71’N, 51 21.59’W) during a period of high river discharge. The collection site (Station 10, 4° 52.71’N, 51° 21.59’W; S = 21.0; T = 29.6°C), located ~ 500 Km to the north of the Amazon River mouth, was characterized by the presence of coastal diatoms in the top 8 m of the water column. Sampling was conducted between 0700 and 0900 local time by gently impeller pumping (modified Rule 1800 submersible sump pump) surface water through 10 m of tygon tubing (3 cm) to the ship's deck where it then flowed through a 156 µm mesh into 20 L carboys. In the lab, cells were partitioned into two size fractions by sequential filtration (using a Masterflex peristaltic pump) of the pre-filtered seawater through a 2.0 µm pore-size, 142 mm diameter polycarbonate (PCTE) membrane filter (Sterlitech Corporation, Kent, CWA) and a 0.22 µm pore-size, 142 mm diameter Supor membrane filter (Pall, Port Washington, NY). Metagenomic and non-selective metatranscriptomic analyses were conducted on both pore-size filters; poly(A)-selected (eukaryote-dominated) metatranscriptomic analyses were conducted only on the larger pore-size filter (2.0 µm pore-size). All filters were immediately submerged in RNAlater (Applied Biosystems, Austin, TX) in sterile 50 mL conical tubes, incubated at room temperature overnight and then stored at -80oC until extraction. Filtration and stabilization of each sample was completed within 30 min of water collection. -

Davidge JBC Supplementary.Pdf
promoting access to White Rose research papers Universities of Leeds, Sheffield and York http://eprints.whiterose.ac.uk/ White Rose Research Online URL for this paper: http://eprints.whiterose.ac.uk/7923/ (includes links to Main Article, Supplementary Material and Figures) Published paper Davidge, K.S., Sanguinetti, G., Yee, C.H., Cox, A.G., McLeod, C.W., Monk, C.E., Mann, B.E., Motterlini, R. and Poole, R.K. (2009) Carbon monoxide-releasing antibacterial molecules target respiration and global transcriptional regulators. Journal of Biological Chemistry, 284 (7). pp. 4516-4524. http://dx.doi.org/10.1074/jbc.M808210200 Supplementary Material White Rose Research Online [email protected] Supplementary Material Carbon monoxide-releasing antibacterial molecules target respiration and global transcriptional regulators Kelly S Davidge, Guido Sanguinetti, Chu Hoi Yee, Alan G Cox, Cameron W McLeod, Claire E Monk, Brian E Mann, Roberto Motterlini and Robert K Poole Contents Page Number Supplementary Figure S1 3 Inhibition by CORM-3 of E. coli cultures grown in defined medium anaerobically and aerobically Supplementary Figure S2 4 Viability assays showing survival of anaerobically and aerobically E. coli in defined growth medium Supplementary Figure S3 5 Reaction of terminal oxidases in vivo on addition of RuCl2(DMSO)4 to intact cells in a dual-wavelength spectrophotometer Supplementary Figure S4 6 CORM-3 generates carbonmonoxycytochrome bd in vivo and depresses synthesis of cytochrome bo' Supplementary Figure S5 7 Expression of spy-lacZ activity -

Structure of Soybean Serine Acetyltransferase
THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 288, NO. 51, pp. 36463–36472, December 20, 2013 Published in the U.S.A. Structure of Soybean Serine Acetyltransferase and Formation of the Cysteine Regulatory Complex as a Molecular Chaperone* Received for publication, October 14, 2013, and in revised form, November 4, 2013 Published, JBC Papers in Press, November 13, 2013, DOI 10.1074/jbc.M113.527143 Hankuil Yi‡1, Sanghamitra Dey§1, Sangaralingam Kumaran¶, Soon Goo Leeʈ, Hari B. Krishnan**, and Joseph M. Jezʈ2 From the ‡Department of Biological Sciences, Chungnam National University, 220 Gung-Dong, Yuseong-Gu, Daejeon 305-764,Korea, the §Department of Biological Sciences, Presidency University, Kolkata, West Bengal 700073, India, the ¶Council of Scientific and Industrial Research, Institute of Microbial Technology, Sector 39-A, Chandigarh 160036, India, the ʈDepartment of Biology, Washington University, St. Louis, Missouri 63130, and the **Plant Genetics Research Unit, United States Department of Agriculture- Agricultural Research Service, Department of Agronomy, University of Missouri, Columbia, Missouri 65211 Background: Serine acetyltransferase (SAT) catalyzes the limiting step in cysteine biosynthesis. Results: Analysis of soybean SAT provides insight into catalysis and protein-protein interactions. Downloaded from Conclusion: Key structural features are required for catalysis and formation of a stable macromolecular complex. Significance: A new role for protein complex formation in plant cysteine biosynthesis is proposed. Serine acetyltransferase (SAT) catalyzes the limiting reaction sis plays a central role in fixing inorganic sulfur from the envi- in plant and microbial biosynthesis of cysteine. In addition to its ronment into the metabolic precursor for cellular thiol-con- http://www.jbc.org/ enzymatic function, SAT forms a macromolecular complex with taining compounds (3–6). -

Letters to Nature
letters to nature Received 7 July; accepted 21 September 1998. 26. Tronrud, D. E. Conjugate-direction minimization: an improved method for the re®nement of macromolecules. Acta Crystallogr. A 48, 912±916 (1992). 1. Dalbey, R. E., Lively, M. O., Bron, S. & van Dijl, J. M. The chemistry and enzymology of the type 1 27. Wolfe, P. B., Wickner, W. & Goodman, J. M. Sequence of the leader peptidase gene of Escherichia coli signal peptidases. Protein Sci. 6, 1129±1138 (1997). and the orientation of leader peptidase in the bacterial envelope. J. Biol. Chem. 258, 12073±12080 2. Kuo, D. W. et al. Escherichia coli leader peptidase: production of an active form lacking a requirement (1983). for detergent and development of peptide substrates. Arch. Biochem. Biophys. 303, 274±280 (1993). 28. Kraulis, P.G. Molscript: a program to produce both detailed and schematic plots of protein structures. 3. Tschantz, W. R. et al. Characterization of a soluble, catalytically active form of Escherichia coli leader J. Appl. Crystallogr. 24, 946±950 (1991). peptidase: requirement of detergent or phospholipid for optimal activity. Biochemistry 34, 3935±3941 29. Nicholls, A., Sharp, K. A. & Honig, B. Protein folding and association: insights from the interfacial and (1995). the thermodynamic properties of hydrocarbons. Proteins Struct. Funct. Genet. 11, 281±296 (1991). 4. Allsop, A. E. et al.inAnti-Infectives, Recent Advances in Chemistry and Structure-Activity Relationships 30. Meritt, E. A. & Bacon, D. J. Raster3D: photorealistic molecular graphics. Methods Enzymol. 277, 505± (eds Bently, P. H. & O'Hanlon, P. J.) 61±72 (R. Soc. Chem., Cambridge, 1997). -

Production of L-Carnitine by Secondary Metabolism of Bacteria Vicente Bernal, Ángel Sevilla, Manuel Cánovas and José L Iborra*
Microbial Cell Factories BioMed Central Review Open Access Production of L-carnitine by secondary metabolism of bacteria Vicente Bernal, Ángel Sevilla, Manuel Cánovas and José L Iborra* Address: Department of Biochemistry and Molecular Biology B and Immunology, Campus of Espinardo, University of Murcia, E-30100, Spain Email: Vicente Bernal - [email protected]; Ángel Sevilla - [email protected]; Manuel Cánovas - [email protected]; José L Iborra* - [email protected] * Corresponding author Published: 2 October 2007 Received: 19 July 2007 Accepted: 2 October 2007 Microbial Cell Factories 2007, 6:31 doi:10.1186/1475-2859-6-31 This article is available from: http://www.microbialcellfactories.com/content/6/1/31 © 2007 Bernal et al; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract The increasing commercial demand for L-carnitine has led to a multiplication of efforts to improve its production with bacteria. The use of different cell environments, such as growing, resting, permeabilized, dried, osmotically stressed, freely suspended and immobilized cells, to maintain enzymes sufficiently active for L-carnitine production is discussed in the text. The different cell states of enterobacteria, such as Escherichia coli and Proteus sp., which can be used to produce L- carnitine from crotonobetaine or D-carnitine as substrate, are analyzed. Moreover, the combined application of both bioprocess and metabolic engineering has allowed a deeper understanding of the main factors controlling the production process, such as energy depletion and the alteration of the acetyl-CoA/CoA ratio which are coupled to the end of the biotransformation. -
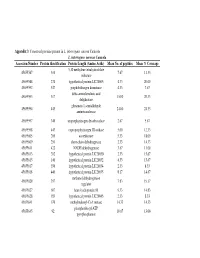
Appendix 3 and 4
Appendix 3 : Conserved proteins present in L. interrogans serovar Canicola L. interrogans serovar Canicola Accession Number Protein identification Protein Length (Amino Acids) Mean No. of peptides Mean % Coverage 5,10 methylene tetrahydrofolate 45655587 310 7.67 11.33 reductase 45655588 274 hypothetical protein LIC20005 4.33 20.00 45655592 547 porphobilinogen deaminase 4.33 7.67 delta-aminolevulinic acid 45655593 317 15.00 20.33 dehydratase glutamate-1-semialdehyde 45655594 443 24.00 24.33 aminotransferase 45655597 340 uroporphyrinogen decarboxylase 2.67 5.67 45655598 443 coproporphyrinogen III oxidase 5.00 12.33 45655605 200 azoreductase 5.33 18.00 45655609 251 short-chain dehydrogenase 2.33 14.33 45655611 422 NADH dehydrogenase 3.67 11.00 45655613 202 hypothetical protein LIC20030 2.33 15.67 45655615 140 hypothetical protein LIC20032 4.33 15.67 45655617 356 hypothetical protein LIC20034 2.33 8.33 45655618 440 hypothetical protein LIC20035 8.17 14.67 methanol dehydrogenase 45655620 357 7.83 19.17 regulator 45655627 607 heat shock protein 90 9.33 14.83 45655628 189 hypothetical protein LIC20045 2.33 8.33 45655641 670 methylmalonyl-CoA mutase 14.33 14.33 phosphoribosyl-ATP 45655645 92 10.67 13.00 pyrophosphatase 3-oxoacyl-(acyl-carrier protein) 45655647 254 4.00 17.00 reductase 45655648 77 acyl carrier protein 3.67 9.00 45655661 171 hypothetical protein LIC20078 5.00 26.00 4-hydroxybenzoyl-CoA 45655663 142 3.33 11.67 thioesterase S-adenosyl-L-homocysteine 45655666 436 13.00 19.67 hydrolase B12-dependent methionine 45655668 1247 42.67 21.67 -

Product Sheet Info
Product Information Sheet for NR-19703 Vibrio cholerae Gateway® Clone Set, Growth Conditions: Media: Recombinant in Escherichia coli, Plate 25 LB broth or agar containing 50 µg/mL kanamycin Incubation: Catalog No. NR-19703 Temperature: E. coli, strain DH10B-T1 clones should be This reagent is the tangible property of the U.S. Government. grown at 37°C. Atmosphere: Aerobic For research use only. Not for human use. Propagation: 1. Scrape top of frozen well with a pipette tip and streak onto Contributor: agar plate. Pathogen Functional Genomics Resource Center at the J. 2. Incubate the plates at 37°C for 1 day. Craig Venter Institute Citation: Manufacturer: Acknowledgment for publications should read “The following BEI Resources reagent was obtained through BEI Resources, NIAID, NIH: ® Vibrio cholerae Gateway Clone Set, Recombinant in Product Description: Escherichia coli, Plate 25, NR-19703.” Production in the 96-well format has increased risk of cross- contamination between adjacent wells. Individual clones Biosafety Level: 1 should be purified (e.g. single colony isolation and purification Appropriate safety procedures should always be used with this using good microbiological practices) and sequence-verified material. Laboratory safety is discussed in the following prior to use. BEI Resources does not confirm or validate publication: U.S. Department of Health and Human Services, individual mutants provided by the contributor. Public Health Service, Centers for Disease Control and Prevention, and National Institutes of Health. Biosafety in The Vibrio cholerae (V. cholerae) Gateway® clone set consists Microbiological and Biomedical Laboratories. 5th ed. of 46 plates which contain 3813 sequence validated clones Washington, DC: U.S.