Expression Profiles of Trna-Derived Fragments and Their Potential Roles in Multiple Myeloma
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

The Three-Dimensional Structure of the Cystic Fibrosis Locus: a Dissertation
University of Massachusetts Medical School eScholarship@UMMS GSBS Dissertations and Theses Graduate School of Biomedical Sciences 2014-11-18 The Three-Dimensional Structure of the Cystic Fibrosis Locus: A Dissertation Emily M. Smith University of Massachusetts Medical School Let us know how access to this document benefits ou.y Follow this and additional works at: https://escholarship.umassmed.edu/gsbs_diss Part of the Genetic Processes Commons, Genomics Commons, and the Structural Biology Commons Repository Citation Smith EM. (2014). The Three-Dimensional Structure of the Cystic Fibrosis Locus: A Dissertation. GSBS Dissertations and Theses. https://doi.org/10.13028/M2SK51. Retrieved from https://escholarship.umassmed.edu/gsbs_diss/744 This material is brought to you by eScholarship@UMMS. It has been accepted for inclusion in GSBS Dissertations and Theses by an authorized administrator of eScholarship@UMMS. For more information, please contact [email protected]. THE THREE-DIMENSIONAL STRUCTURE OF THE CYSTIC FIBROSIS LOCUS A Dissertation Presented By EMILY MALINDA SMITH Submitted to the Faculty of the University of Massachusetts Graduate School of Biomedical Sciences, Worcester in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY NOVEMBER 18, 2014 BIOMEDICAL SCIENCES THE THREE-DIMENSIONAL STRUCTURE OF THE CYSTIC FIBROSIS LOCUS Dissertation Presented By Emily Malinda Smith The signatures of the ee signify completion and approval as t the Dissertation The signature of the Chair of the Committee signifies that the written dissertation meets the requirements of the Dissertation Committee Sean omm1ttee The signature of the Dean of the Graduate School of Biomedical Sciences signifies that the student has met al aduation requirements of the school. -

Table SII. Significantly Differentially Expressed Mrnas of GSE23558 Data Series with the Criteria of Adjusted P<0.05 And
Table SII. Significantly differentially expressed mRNAs of GSE23558 data series with the criteria of adjusted P<0.05 and logFC>1.5. Probe ID Adjusted P-value logFC Gene symbol Gene title A_23_P157793 1.52x10-5 6.91 CA9 carbonic anhydrase 9 A_23_P161698 1.14x10-4 5.86 MMP3 matrix metallopeptidase 3 A_23_P25150 1.49x10-9 5.67 HOXC9 homeobox C9 A_23_P13094 3.26x10-4 5.56 MMP10 matrix metallopeptidase 10 A_23_P48570 2.36x10-5 5.48 DHRS2 dehydrogenase A_23_P125278 3.03x10-3 5.40 CXCL11 C-X-C motif chemokine ligand 11 A_23_P321501 1.63x10-5 5.38 DHRS2 dehydrogenase A_23_P431388 2.27x10-6 5.33 SPOCD1 SPOC domain containing 1 A_24_P20607 5.13x10-4 5.32 CXCL11 C-X-C motif chemokine ligand 11 A_24_P11061 3.70x10-3 5.30 CSAG1 chondrosarcoma associated gene 1 A_23_P87700 1.03x10-4 5.25 MFAP5 microfibrillar associated protein 5 A_23_P150979 1.81x10-2 5.25 MUCL1 mucin like 1 A_23_P1691 2.71x10-8 5.12 MMP1 matrix metallopeptidase 1 A_23_P350005 2.53x10-4 5.12 TRIML2 tripartite motif family like 2 A_24_P303091 1.23x10-3 4.99 CXCL10 C-X-C motif chemokine ligand 10 A_24_P923612 1.60x10-5 4.95 PTHLH parathyroid hormone like hormone A_23_P7313 6.03x10-5 4.94 SPP1 secreted phosphoprotein 1 A_23_P122924 2.45x10-8 4.93 INHBA inhibin A subunit A_32_P155460 6.56x10-3 4.91 PICSAR P38 inhibited cutaneous squamous cell carcinoma associated lincRNA A_24_P686965 8.75x10-7 4.82 SH2D5 SH2 domain containing 5 A_23_P105475 7.74x10-3 4.70 SLCO1B3 solute carrier organic anion transporter family member 1B3 A_24_P85099 4.82x10-5 4.67 HMGA2 high mobility group AT-hook 2 A_24_P101651 -

Supplementary Table 1
Supplementary Table 1. 492 genes are unique to 0 h post-heat timepoint. The name, p-value, fold change, location and family of each gene are indicated. Genes were filtered for an absolute value log2 ration 1.5 and a significance value of p ≤ 0.05. Symbol p-value Log Gene Name Location Family Ratio ABCA13 1.87E-02 3.292 ATP-binding cassette, sub-family unknown transporter A (ABC1), member 13 ABCB1 1.93E-02 −1.819 ATP-binding cassette, sub-family Plasma transporter B (MDR/TAP), member 1 Membrane ABCC3 2.83E-02 2.016 ATP-binding cassette, sub-family Plasma transporter C (CFTR/MRP), member 3 Membrane ABHD6 7.79E-03 −2.717 abhydrolase domain containing 6 Cytoplasm enzyme ACAT1 4.10E-02 3.009 acetyl-CoA acetyltransferase 1 Cytoplasm enzyme ACBD4 2.66E-03 1.722 acyl-CoA binding domain unknown other containing 4 ACSL5 1.86E-02 −2.876 acyl-CoA synthetase long-chain Cytoplasm enzyme family member 5 ADAM23 3.33E-02 −3.008 ADAM metallopeptidase domain Plasma peptidase 23 Membrane ADAM29 5.58E-03 3.463 ADAM metallopeptidase domain Plasma peptidase 29 Membrane ADAMTS17 2.67E-04 3.051 ADAM metallopeptidase with Extracellular other thrombospondin type 1 motif, 17 Space ADCYAP1R1 1.20E-02 1.848 adenylate cyclase activating Plasma G-protein polypeptide 1 (pituitary) receptor Membrane coupled type I receptor ADH6 (includes 4.02E-02 −1.845 alcohol dehydrogenase 6 (class Cytoplasm enzyme EG:130) V) AHSA2 1.54E-04 −1.6 AHA1, activator of heat shock unknown other 90kDa protein ATPase homolog 2 (yeast) AK5 3.32E-02 1.658 adenylate kinase 5 Cytoplasm kinase AK7 -

SUPPLEMENTAL TABLE 1: List of Shrnas Used for Validation
Singleton, Kim, Hinz, Marek, Casas-Selves, Hatheway, Tan, DeGregori and Heasley. A Receptor Tyrosine Kinase Network Comprised of FGFRs, EGFR, ERBB2 and MET Drives Growth and Survival of Head and Neck Squamous Carcinoma Cell Lines. MOLPHARM #084111 SUPPLEMENTAL TABLE 1: List of shRNAs used for validation. Singleton, Kim, Hinz, Marek, Casas-Selves, Hatheway, Tan, DeGregori and Heasley. A Receptor Tyrosine Kinase Network Comprised of FGFRs, EGFR, ERBB2 and MET Drives Growth and Survival of Head and Neck Squamous Carcinoma Cell Lines. MOLPHARM #084111 SUPPLEMENTAL TABLE 2: SLATs (E-value cut off of E value ≤2) in UMSCC25, 584-A2, and CCL30. UMSCC25 Rank Gene Symbol p-value [p(wZ)] E value Rank Gene Symbol p-value [p(wZ)] E value 1 CLUAP1 5.15E-07 5.15E-07 48 FHL1 4.84E-04 2.32E-02 2 ZFR 8.78E-07 1.76E-06 49 RABGGTB 5.03E-04 2.46E-02 3 MET 1.94E-06 5.83E-06 50 THPO 5.28E-04 2.64E-02 4 MEMO1 1.95E-06 7.79E-06 50 BCAP29 5.28E-04 2.64E-02 5 NPAT 3.00E-06 1.50E-05 52 KBTBD4 5.28E-04 2.75E-02 6 EPB41L4B 3.18E-06 1.91E-05 53 MEX3B 5.28E-04 2.80E-02 7 G2E3 3.43E-06 2.40E-05 54 EML4 5.29E-04 2.85E-02 8 PSMD13 4.10E-06 3.28E-05 55 C20orf7 5.29E-04 2.91E-02 9 MUC7 5.60E-06 5.04E-05 56 GPR126 5.29E-04 2.96E-02 10 ARSD 5.67E-06 5.67E-05 56 PNPO 5.29E-04 2.96E-02 11 TMEM163 1.13E-05 1.24E-04 58 TJAP1 5.29E-04 3.07E-02 12 PCTK2 1.38E-05 1.65E-04 58 SLC26A2 5.29E-04 3.07E-02 13 ANP32E 1.38E-05 1.80E-04 58 NETO2 5.29E-04 3.07E-02 14 ITGA6 2.62E-05 3.66E-04 61 MAML3 5.29E-04 3.23E-02 15 C14orf147 2.74E-05 4.11E-04 62 HLA-DRB3 5.30E-04 3.29E-02 16 MDH1B 3.18E-05 -
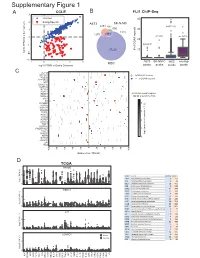
Supplementary Figure 1
6XSSOHPHQWDU\)LJXUH CCLE B FLI1 ChIP-Seq A 3 FRPPRQ 40 (ZLQJ6SHFLILF 2 A673 6.10& 477 Q 690 30 0 UHSHDWV Q Q A 20 í Q 57226 RI**$ ORJ)3.0LQRWKHUFDQFHUV í í A673 6.10& 06& RYHUODS í 0 23 06& ORJ)3.0LQ(ZLQJ6DUFRPD SHDNV SHDNV SHDNV SHDNV & ./) 67($3 (:6)/,ERXQG 3'(% ! **$AUHSHDWV 3335$ ABI3 '1$-& '863 13<5 .&1( 5%0 DOO**$AUHSHDWUHJLRQV 1.; NEDURXQGWKH766 9$9 ;* .&1$% 4 .&1$ 51) 0<20 3 355/ 355T4 &'$ $'5% 2 '&'& 711, 8*7$ 3+263+2 )(=) *1*7 ORJ QXPEHURIUHSHDWV A571 0 13<5 /2;+' /,3, 5$; 0 40 60 20 GLVWDQFHIURP766 NE D TCGA 35$0( 5 code cancer normal tumor LAML Acute Myeloid Leukemia 0 151 ORJ )3.0 ACC AdrenocoƌƟcal carcinoma 0 79 0 BLCA Bladder Urothelial Carcinoma 19 411 LGG Brain Lower Grade Glioma 0 529 COAD Colon adenocarcinoma 41 464 5%0 ESCA Esophageal carcinoma 11 162 GBM Glioblastoma muůƟforme 5 168 5 KICH Kidney Chromophobe 24 65 KIRP Kidney renal papillary cell carcinoma 32 289 LIHC Liver hepatocellular carcinoma 50 374 ORJ )3.0 LUAD Lung adenocarcinoma 59 526 0 LUSC Lung squamous cell carcinoma 49 501 DLBC Lymphoid Neoplasm Diīuse Large B-cell Lym 042 /,3, MESO Mesothelioma 0 1 OV Ovarian serous cystadenocarcinoma 0 379 PAAD PancreĂƟc adenocarcinoma 4 178 5 PCPG Pheochromocytoma and Paraganglioma 3 150 PRAD Prostate adenocarcinoma 52 499 READ Rectum adenocarcinoma 1 6 ORJ )3.0 SARC Sarcoma 0 35 0 SKCM Skin Cutaneous Melanoma 1 471 STAD Stomach adenocarcinoma 32 375 /2;+' WXPRU TGCT dĞƐƟĐƵůĂƌ'ĞƌŵĞůůdƵŵŽƌƐ 0156 THCA Thyroid carcinoma 58 510 QRUPDO UCS Uterine Carcinosarcoma 0 56 5 ORJ )3.0 0 / / 29 $&& 8&6 /** *%0 -

Inhibition of HIV-1 Gene Transcription by KAP1 in Myeloid Lineage
www.nature.com/scientificreports OPEN Inhibition of HIV‑1 gene transcription by KAP1 in myeloid lineage Amina Ait‑Ammar1,2,3, Maxime Bellefroid3, Fadoua Daouad1, Valérie Martinelli4, Jeanne Van Assche1, Clémentine Wallet1, Anthony Rodari3, Marco De Rovere1, Birthe Fahrenkrog4, Christian Schwartz1, Carine Van Lint3,5*, Virginie Gautier2,5* & Olivier Rohr1,5* HIV‑1 latency generates reservoirs that prevent viral eradication by the current therapies. To fnd strategies toward an HIV cure, detailed understandings of the molecular mechanisms underlying establishment and persistence of the reservoirs are needed. The cellular transcription factor KAP1 is known as a potent repressor of gene transcription. Here we report that KAP1 represses HIV‑1 gene expression in myeloid cells including microglial cells, the major reservoir of the central nervous system. Mechanistically, KAP1 interacts and colocalizes with the viral transactivator Tat to promote its degradation via the proteasome pathway and repress HIV‑1 gene expression. In myeloid models of latent HIV‑1 infection, the depletion of KAP1 increased viral gene elongation and reactivated HIV‑1 expression. Bound to the latent HIV‑1 promoter, KAP1 associates and cooperates with CTIP2, a key epigenetic silencer of HIV‑1 expression in microglial cells. In addition, Tat and CTIP2 compete for KAP1 binding suggesting a dynamic modulation of the KAP1 cellular partners upon HIV‑1 infection. Altogether, our results suggest that KAP1 contributes to the establishment and the persistence of HIV‑1 latency in myeloid cells. Te pandemic of HIV-1 infections is a global health problem. Current cART (combination antiretroviral therapy) efciently suppresses viral expression below clinical detection levels but fails to eradicate latently infected res- ervoirs, the main obstacles toward an HIV cure. -

CRISPR-Based Technology to Silence the Expression of Incrnas COMMENTARY James D
COMMENTARY CRISPR-based technology to silence the expression of IncRNAs COMMENTARY James D. Phelana and Louis M. Staudta,1 The human genome contains more than 3 billion base by disrupting noncoding regulatory elements (13). pairs, and some estimates suggest that nearly 75% of However, CRISPR screens with Cas9 are less able to the genome may be transcribed (1), yet only a small reliably elucidate lncRNA function, as focal indels may fraction (1 to 2%) of the genome that encodes for pro- not perturb lncRNA function, and the functional do- tein coding regions has been systematically probed mains of most lncRNAs have not been mapped. for function. The transcribed genome consists of both Previous studies have shown that Cas9 can be short noncoding RNAs (ncRNAs) and long ncRNAs engineered to silence gene expression (rather than (lncRNAs). The lncRNAs are defined as being greater induce double-stranded breaks) by fusing transcrip- than 200 nucleotides in length and may be capped, tional repressor domains to an enzymatically inactive polyadenylated, and spliced like protein-coding genes. “dead” Cas9 (dCas9) isoform (14). In these CRISPR By definition, lncRNAs do not contain open reading interference (CRISPRi) systems, a single guide RNA framesofgreaterthan50aminoacids,butrecentwork (sgRNA) is used to recruit dCas9 repressor domain fu- has uncovered a new world of smaller “microproteins” sions to a specific genomic location where they can that are encoded by lncRNAs (2). Other functions of repress the transcription. Raffeiner et al. (7) turn to lncRNAs include their ability to form aptamers, which this technology to probe the function of lncRNAs that bind and regulate protein complexes (3). -
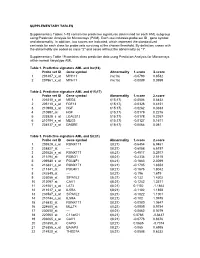
Supplementary Tables 1-18 Contain the Predictive Signatures Determined for Each AML Subgroup Using Prediction Analysis for Microarrays (PAM)
SUPPLEMENTARY TABLES Supplementary Tables 1-18 contain the predictive signatures determined for each AML subgroup using Prediction Analysis for Microarrays (PAM). Each row indicates probe set ID , gene symbol and abnormality. In addition, two scores are indicated, which represent the standardized centroids for each class for probe sets surviving at the chosen threshold. By definition, cases with the abnormality are coded as class "2" and cases without the abnormality as "1". Supplementary Table 19 contains class prediction data using Prediction Analysis for Microarrays within normal karyotype AML. Table 1. Predictive signature AML and inv(16) Probe set ID Gene symbol Abnormality 1.score 2.score 1 201497_x_at MYH11 inv(16) -0.0793 0.8532 2 207961_x_at MYH11 inv(16) -0.0089 0.0959 Table 2. Predictive signature AML and t(15;17) Probe set ID Gene symbol Abnormality 1.score 2.score 1 226210_s_at MEG3 t(15;17) -0.0506 0.6432 2 205110_s_at FGF13 t(15;17) -0.0326 0.4151 3 210998_s_at HGF t(15;17) -0.0262 0.3333 4 210997_at HGF t(15;17) -0.0179 0.2276 5 223828_s_at LGALS12 t(15;17) -0.0178 0.2267 6 210794_s_at MEG3 t(15;17) -0.0127 0.1611 7 204537_s_at GABRE t(15;17) -0.0064 0.081 Table 3. Predictive signature AML and t(8;21) Probe set ID Gene symbol Abnormality 1.score 2.score 1 205529_s_at RUNX1T1 t(8;21) -0.6454 6.9461 2 228827_at --- t(8;21) -0.6058 6.5197 3 205528_s_at RUNX1T1 t(8;21) -0.4917 5.2917 4 213194_at ROBO1 t(8;21) -0.2334 2.5115 5 206940_s_at POU4F1 t(8;21) -0.1883 2.0269 6 216831_s_at RUNX1T1 t(8;21) -0.1705 1.8353 7 211341_at -

Clinical Efficacy and Immune Regulation with Peanut Oral
Clinical efficacy and immune regulation with peanut oral immunotherapy Stacie M. Jones, MD,a Laurent Pons, PhD,b Joseph L. Roberts, MD, PhD,b Amy M. Scurlock, MD,a Tamara T. Perry, MD,a Mike Kulis, PhD,b Wayne G. Shreffler, MD, PhD,c Pamela Steele, CPNP,b Karen A. Henry, RN,a Margaret Adair, MD,b James M. Francis, PhD,d Stephen Durham, MD,d Brian P. Vickery, MD,b Xiaoping Zhong, MD, PhD,b and A. Wesley Burks, MDb Little Rock, Ark, Durham, NC, New York, NY, and London, United Kingdom Background: Oral immunotherapy (OIT) has been thought to noted during OIT resolved spontaneously or with induce clinical desensitization to allergenic foods, but trials antihistamines. By 6 months, titrated skin prick tests and coupling the clinical response and immunologic effects of peanut activation of basophils significantly declined. Peanut-specific OIT have not been reported. IgE decreased by 12 to 18 months, whereas IgG4 increased Objective: The study objective was to investigate the clinical significantly. Serum factors inhibited IgE–peanut complex efficacy and immunologic changes associated with OIT. formation in an IgE-facilitated allergen binding assay. Secretion Methods: Children with peanut allergy underwent an OIT of IL-10, IL-5, IFN-g, and TNF-a from PBMCs increased over protocol including initial day escalation, buildup, and a period of 6 to 12 months. Peanut-specific forkhead box protein maintenance phases, and then oral food challenge. Clinical 3 T cells increased until 12 months and decreased thereafter. In response and immunologic changes were evaluated. addition, T-cell microarrays showed downregulation of genes in Results: Of 29 subjects who completed the protocol, 27 ingested apoptotic pathways. -

Supplementary Figure S1 A
Supplementary Figure S1 A B C 1 4 1 3 1 2 1 2 1 0 1 1 (Normalized signal values)] 2 (Normalized signal values) (Normalized 8 2 10 Log 6 AFFX-BioC AFFX-BioB AFFX-BioDn AFFX-CreX hDFs [Log 6 8 10 12 14 hOFs [Log2 (Normalized signal values)] Supplementary Figure S2 GLYCOLYSIS PENTOSE-PHOSPHATE PATHWAY Glucose Purine/pyrimidine Glucose-6-phosphate metabolism AMINO ACID Fluctose-6-phosphate AMPK METABOLISM TIGAR PFKFB2 methylgloxal GloI Ser, Gly, Thr Glyceraldehyde-3-phosphate ALDH Lactate PYRUVATE LDH METABOLISM acetic acid Ethanol Pyruvate GLYCOSPHINGOLIPID NADH BIOSYNTHESIS Ala, Cys DLD PDH PDK3 DLAT Fatty acid Lys, Trp, Leu, Acetyl CoA ACAT2 Ile, Tyr, Phe β-OXIDATION ACACA Citrate Asp, Asn Citrate Acetyl CoA Oxaloacetate Isocitrate MDH1 IDH1 Glu, Gln, His, ME2 TCA Pro, Arg 2-Oxoglutarate MDH1 CYCLE Pyruvate Malate ME2 GLUTAMINOLYSIS FH Succinyl-CoA Fumalate SUCLA2 Tyr, Phe Var, Ile, Met Supplementary Figure S3 Entrez Gene Symbol Gene Name hODs hDFs hOF-iPSCs GeneID 644 BLVRA biliverdin reductase A 223.9 259.3 253.0 3162 HMOX1 heme oxygenase 1 1474.2 2698.0 452.3 9365 KL klotho 54.1 44.8 36.5 nicotinamide 10135 NAMPT 827.7 626.2 2999.8 phosphoribosyltransferase nuclear factor (erythroid- 4780 NFE2L2 2134.5 1331.7 1006.2 derived 2) related factor 2 peroxisome proliferator- 5467 PPARD 1534.6 1352.9 330.8 activated receptor delta peroxisome proliferator- 5468 PPARG 524.4 100.8 63.0 activated receptor gamma 5621 PRNP prion protein 4059.0 3134.1 1065.5 5925 RB1 retinoblastoma 1 882.9 805.8 739.3 23411 SIRT1 sirtuin 1 231.5 216.8 1676.0 7157 TP53