Reverse Engineering of Ewing Sarcoma Regulatory Network Uncovers PAX7 and RUNX3 As Master Regulators Associated with Good Prognosis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Aberrant Methylation Underlies Insulin Gene Expression in Human Insulinoma
ARTICLE https://doi.org/10.1038/s41467-020-18839-1 OPEN Aberrant methylation underlies insulin gene expression in human insulinoma Esra Karakose1,6, Huan Wang 2,6, William Inabnet1, Rajesh V. Thakker 3, Steven Libutti4, Gustavo Fernandez-Ranvier 1, Hyunsuk Suh1, Mark Stevenson 3, Yayoi Kinoshita1, Michael Donovan1, Yevgeniy Antipin1,2, Yan Li5, Xiaoxiao Liu 5, Fulai Jin 5, Peng Wang 1, Andrew Uzilov 1,2, ✉ Carmen Argmann 1, Eric E. Schadt 1,2, Andrew F. Stewart 1,7 , Donald K. Scott 1,7 & Luca Lambertini 1,6 1234567890():,; Human insulinomas are rare, benign, slowly proliferating, insulin-producing beta cell tumors that provide a molecular “recipe” or “roadmap” for pathways that control human beta cell regeneration. An earlier study revealed abnormal methylation in the imprinted p15.5-p15.4 region of chromosome 11, known to be abnormally methylated in another disorder of expanded beta cell mass and function: the focal variant of congenital hyperinsulinism. Here, we compare deep DNA methylome sequencing on 19 human insulinomas, and five sets of normal beta cells. We find a remarkably consistent, abnormal methylation pattern in insu- linomas. The findings suggest that abnormal insulin (INS) promoter methylation and altered transcription factor expression create alternative drivers of INS expression, replacing cano- nical PDX1-driven beta cell specification with a pathological, looping, distal enhancer-based form of transcriptional regulation. Finally, NFaT transcription factors, rather than the cano- nical PDX1 enhancer complex, are predicted to drive INS transactivation. 1 From the Diabetes Obesity and Metabolism Institute, The Department of Surgery, The Department of Pathology, The Department of Genetics and Genomics Sciences and The Institute for Genomics and Multiscale Biology, The Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA. -
Nucleoporin 107, 62 and 153 Mediate Kcnq1ot1 Imprinted Domain Regulation in Extraembryonic Endoderm Stem Cells
ARTICLE DOI: 10.1038/s41467-018-05208-2 OPEN Nucleoporin 107, 62 and 153 mediate Kcnq1ot1 imprinted domain regulation in extraembryonic endoderm stem cells Saqib S. Sachani 1,2,3,4, Lauren S. Landschoot1,2, Liyue Zhang1,2, Carlee R. White1,2, William A. MacDonald3,4, Michael C. Golding 5 & Mellissa R.W. Mann 3,4 1234567890():,; Genomic imprinting is a phenomenon that restricts transcription to predominantly one par- ental allele. How this transcriptional duality is regulated is poorly understood. Here we perform an RNA interference screen for epigenetic factors involved in paternal allelic silen- cing at the Kcnq1ot1 imprinted domain in mouse extraembryonic endoderm stem cells. Multiple factors are identified, including nucleoporin 107 (NUP107). To determine NUP107’s role and specificity in Kcnq1ot1 imprinted domain regulation, we deplete Nup107, as well as Nup62, Nup98/96 and Nup153. Nup107, Nup62 and Nup153, but not Nup98/96 depletion, reduce Kcnq1ot1 noncoding RNA volume, displace the Kcnq1ot1 domain from the nuclear periphery, reactivate a subset of normally silent paternal alleles in the domain, alter histone modifications with concomitant changes in KMT2A, EZH2 and EHMT2 occupancy, as well as reduce cohesin interactions at the Kcnq1ot1 imprinting control region. Our results establish an important role for specific nucleoporins in mediating Kcnq1ot1 imprinted domain regulation. 1 Departments of Obstetrics & Gynaecology, and Biochemistry, Western University, Schulich School of Medicine and Dentistry, London, ON N6A 5W9, Canada. 2 Children’s Health Research Institute, London, ON N6C 2V5, Canada. 3 Departments of Obstetrics, Gynecology and Reproductive Sciences, University of Pittsburgh School of Medicine, Pittsburgh, PA 15213, USA. 4 Magee-Womens Research Institute, Pittsburgh, PA 15213, USA. -
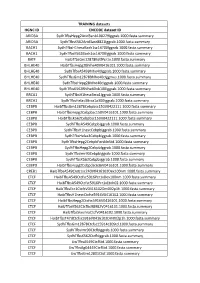
TRAINING Datasets HGNC ID ENCODE Dataset ID ARID3A
TRAINING datasets HGNC ID ENCODE dataset ID ARID3A SydhT+sHepg2Arid3anb100279Iggrab.1000.fasta.summary ARID3A SydhT+sK562Arid3asC8821Iggrab.1000.fasta.summary BACH1 SydhT+sH1hesCBaCh1sC14700Iggrab.1000.fasta.summary BACH1 SydhT+sK562BaCh1sC14700Iggrab.1000.fasta.summary BATF HaibT+sGm12878BaJPCr1x.1000.fasta.summary BHLHE40 HaibT+sHepg2Bhlhe40V0416101.1000.fasta.summary BHLHE40 SydhT+sA549Bhlhe40Iggrab.1000.fasta.summary BHLHE40 SydhT+sGm12878Bhlhe40CIggmus.1000.fasta.summary BHLHE40 SydhT+sHepg2Bhlhe40CIggrab.1000.fasta.summary BHLHE40 SydhT+sK562Bhlhe40nb100Iggrab.1000.fasta.summary BRCA1 SydhT+sH1hesCBrCa1Iggrab.1000.fasta.summary BRCA1 SydhT+sHelas3BrCa1a300Iggrab.1000.fasta.summary CEBPB HaibT+sGm12878CebpbsC150V0422111.1000.fasta.summary CEBPB HaibT+sHepg2CebpbsC150V0416101.1000.fasta.summary CEBPB HaibT+sK562CebpbsC150V0422111.1000.fasta.summary CEBPB SydhT+sA549CebpbIggrab.1000.fasta.summary CEBPB SydhT+sH1hesCCebpbIggrab.1000.fasta.summary CEBPB SydhT+sHelas3CebpbIggrab.1000.fasta.summary CEBPB SydhT+sHepg2CebpbForsklnStd.1000.fasta.summary CEBPB SydhT+sHepg2CebpbIggrab.1000.fasta.summary CEBPB SydhT+sImr90CebpbIggrab.1000.fasta.summary CEBPB SydhT+sK562CebpbIggrab.1000.fasta.summary CEBPD HaibT+sHepg2CebpdsC636V0416101.1000.fasta.summary CREB1 HaibT+sA549Creb1sC240V0416102Dex100nm.1000.fasta.summary CTCF HaibT+sA549CtCfsC5916PCr1xDex100nm.1000.fasta.summary CTCF HaibT+sA549CtCfsC5916PCr1xEtoh02.1000.fasta.summary CTCF HaibT+sECC1CtCfCV0416102Dm002p1h.1000.fasta.summary CTCF HaibT+sH1hesCCtCfsC5916V0416102.1000.fasta.summary -

1 the Neuroprotective Transcription Factor ATF5 Is Decreased And
The neuroprotective transcription factor ATF5 is decreased and sequestered into polyglutamine inclusions in Huntington’s disease Ivó H. Hernández1,2,3, Jesús Torres-Peraza1,2,5, María Santos-Galindo1,2, Eloísa Ramos-Morón4, María R. Fernández-Fernández1,2, María J. Pérez-Álvarez1,2,3, Antonio Miranda-Vizuete4 and José J. Lucas1,2* 1 Centro de Biología Molecular Severo Ochoa (CBM”SO”) CSIC/UAM, Madrid, Spain. 2 Instituto de Salud Carlos III, Networking Research Center on Neurodegenerative Diseases (CIBERNED), Spain. 3 Departamento de Biología, Facultad de Ciencias, Universidad Autónoma de Madrid, Madrid, Spain. 4 Instituto de Biomedicina de Sevilla, Hospital Universitario Virgen del Rocío/CSIC/Universidad de Sevilla, 41013 Sevilla, Spain 5 Present Address: Gerència d’Atenció Primària del Servei de Salut de les Illes Balears (IB-SALUT), Palma, Spain *Corresponding author: José J. Lucas [email protected] http://www.cbm.uam.es/lineas/lucasgroup.htm provided by Digital.CSIC View metadata, citation and similar papers at core.ac.uk CORE brought to you by 1 Abstract Activating transcription factor-5 (ATF5) is a stress-response transcription factor induced upon different cell stressors like fasting, amino-acid limitation, cadmium or arsenite. ATF5 is also induced, and promotes transcription of anti-apoptotic target genes like MCL1, during the unfolded protein response (UPR) triggered by endoplasmic reticulum stress. In the brain, high ATF5 levels are found in gliomas and also in neural progenitor cells, which need to decrease their ATF5 levels for differentiation into mature neurons or glia. This initially led to believe that ATF5 is not expressed in adult neurons. More recently, we reported basal neuronal ATF5 expression in adult mouse brain and its neuroprotective induction during UPR in a mouse model of status epilepticus. -

Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model
Downloaded from http://www.jimmunol.org/ by guest on September 25, 2021 T + is online at: average * The Journal of Immunology , 34 of which you can access for free at: 2016; 197:1477-1488; Prepublished online 1 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600589 http://www.jimmunol.org/content/197/4/1477 Molecular Profile of Tumor-Specific CD8 Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. Waugh, Sonia M. Leach, Brandon L. Moore, Tullia C. Bruno, Jonathan D. Buhrman and Jill E. Slansky J Immunol cites 95 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2016/07/01/jimmunol.160058 9.DCSupplemental This article http://www.jimmunol.org/content/197/4/1477.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 25, 2021. The Journal of Immunology Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. -

Jmjd2c Facilitates the Assembly of Essential Enhancer-Protein Complexes at the Onset of Embryonic Stem Cell Differentiation Rute A
© 2017. Published by The Company of Biologists Ltd | Development (2017) 144, 567-579 doi:10.1242/dev.142489 STEM CELLS AND REGENERATION RESEARCH ARTICLE Jmjd2c facilitates the assembly of essential enhancer-protein complexes at the onset of embryonic stem cell differentiation Rute A. Tomaz1,JenniferL.Harman2, Donja Karimlou1, Lauren Weavers1, Lauriane Fritsch3, Tony Bou-Kheir1, Emma Bell1, Ignacio del Valle Torres4, Kathy K. Niakan4,CynthiaFisher5, Onkar Joshi6, Hendrik G. Stunnenberg6, Edward Curry1, Slimane Ait-Si-Ali3, Helle F. Jørgensen2 and Véronique Azuara1,* ABSTRACT implantation, a second extra-embryonic lineage, the primitive Jmjd2 H3K9 demethylases cooperate in promoting mouse embryonic endoderm, emerges at the ICM surface. Concurrently, the ICM stem cell (ESC) identity. However, little is known about their maintains its pluripotency as it matures into the epiblast but importance at the exit of ESC pluripotency. Here, we reveal that ultimately goes on to form the three primary germ layers and germ Jmjd2c facilitates this process by stabilising the assembly of cells upon gastrulation (Boroviak and Nichols, 2014; Rossant, mediator-cohesin complexes at lineage-specific enhancers. 2008). Functionally, we show that Jmjd2c is required in ESCs to initiate Pluripotent mouse embryonic stem cells (ESCs) are derived from appropriate gene expression programs upon somatic multi-lineage ICM cells, and can self-renew and faithfully maintain an differentiation. In the absence of Jmjd2c, differentiation is stalled at an undifferentiated state in vitro in the presence of leukaemia inhibitory early post-implantation epiblast-like stage, while Jmjd2c-knockout factor (LIF) and serum components, while preserving their multi- ESCs remain capable of forming extra-embryonic endoderm lineage differentiation capacity (Evans and Kaufman, 1981; Martin, derivatives. -

Protein Interaction Network of Alternatively Spliced Isoforms from Brain Links Genetic Risk Factors for Autism
ARTICLE Received 24 Aug 2013 | Accepted 14 Mar 2014 | Published 11 Apr 2014 DOI: 10.1038/ncomms4650 OPEN Protein interaction network of alternatively spliced isoforms from brain links genetic risk factors for autism Roser Corominas1,*, Xinping Yang2,3,*, Guan Ning Lin1,*, Shuli Kang1,*, Yun Shen2,3, Lila Ghamsari2,3,w, Martin Broly2,3, Maria Rodriguez2,3, Stanley Tam2,3, Shelly A. Trigg2,3,w, Changyu Fan2,3, Song Yi2,3, Murat Tasan4, Irma Lemmens5, Xingyan Kuang6, Nan Zhao6, Dheeraj Malhotra7, Jacob J. Michaelson7,w, Vladimir Vacic8, Michael A. Calderwood2,3, Frederick P. Roth2,3,4, Jan Tavernier5, Steve Horvath9, Kourosh Salehi-Ashtiani2,3,w, Dmitry Korkin6, Jonathan Sebat7, David E. Hill2,3, Tong Hao2,3, Marc Vidal2,3 & Lilia M. Iakoucheva1 Increased risk for autism spectrum disorders (ASD) is attributed to hundreds of genetic loci. The convergence of ASD variants have been investigated using various approaches, including protein interactions extracted from the published literature. However, these datasets are frequently incomplete, carry biases and are limited to interactions of a single splicing isoform, which may not be expressed in the disease-relevant tissue. Here we introduce a new interactome mapping approach by experimentally identifying interactions between brain-expressed alternatively spliced variants of ASD risk factors. The Autism Spliceform Interaction Network reveals that almost half of the detected interactions and about 30% of the newly identified interacting partners represent contribution from splicing variants, emphasizing the importance of isoform networks. Isoform interactions greatly contribute to establishing direct physical connections between proteins from the de novo autism CNVs. Our findings demonstrate the critical role of spliceform networks for translating genetic knowledge into a better understanding of human diseases. -

Feedback Regulation Between Initiation and Maturation Networks Orchestrates the Chromatin Dynamics of Epidermal Lineage
bioRxiv preprint doi: https://doi.org/10.1101/349308; this version posted June 18, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Li et al., p. 1 Feedback Regulation between Initiation and Maturation Networks Orchestrates the Chromatin Dynamics of Epidermal Lineage Commitment Lingjie Li1,3,4, Yong Wang2,4,7,8*, Jessica L. Torkelson1,3*, Gautam Shankar1, Jillian M. Pattison1,3, Hanson H. Zhen1,3, Zhana Duren2,4,7, Fengqin Fang5, Sandra P. Melo1, Samantha N. Piekos1,3, Jiang Li1, Eric J. Liaw1, Lang Chen7, Rui Li1,4, Marius Wernig6, Wing H. Wong2,4, Howard Y. Chang1,4, Anthony E. Oro1,3,9 1 Program in Epithelial Biology and Department of Dermatology 2 Department of Statistics and Biomedical Data Science 3 Center for Definitive and Curative Medicine 4 Center for Personal Dynamic Regulome 5 Division of Immunology and Rheumatology, Department of Medicine, 6 Institute for Stem Cell Biology and Regenerative Medicine, Department of Pathology, Stanford University School of Medicine, Stanford, CA 94305, USA. 7 CEMS, NCMIS, MDIS, Academy of Mathematics & Systems Science, Chinese Academy of Sciences, Beijing,100080, China 8 Center for Excellence in Animal Evolution and Genetics, Chinese Academy of Sciences, Kunming, 650223, China *These authors made equal and independent contributions. 9 Correspondence to Lead Contact: Anthony E. Oro at [email protected] bioRxiv preprint doi: https://doi.org/10.1101/349308; this version posted June 18, 2018. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

A Clinicopathological and Molecular Genetic Analysis of Low-Grade Glioma in Adults
A CLINICOPATHOLOGICAL AND MOLECULAR GENETIC ANALYSIS OF LOW-GRADE GLIOMA IN ADULTS Presented by ANUSHREE SINGH MSc A thesis submitted in partial fulfilment of the requirements of the University of Wolverhampton for the degree of Doctor of Philosophy Brain Tumour Research Centre Research Institute in Healthcare Sciences Faculty of Science and Engineering University of Wolverhampton November 2014 i DECLARATION This work or any part thereof has not previously been presented in any form to the University or to any other body whether for the purposes of assessment, publication or for any other purpose (unless otherwise indicated). Save for any express acknowledgments, references and/or bibliographies cited in the work, I confirm that the intellectual content of the work is the result of my own efforts and of no other person. The right of Anushree Singh to be identified as author of this work is asserted in accordance with ss.77 and 78 of the Copyright, Designs and Patents Act 1988. At this date copyright is owned by the author. Signature: Anushree Date: 30th November 2014 ii ABSTRACT The aim of the study was to identify molecular markers that can determine progression of low grade glioma. This was done using various approaches such as IDH1 and IDH2 mutation analysis, MGMT methylation analysis, copy number analysis using array comparative genomic hybridisation and identification of differentially expressed miRNAs using miRNA microarray analysis. IDH1 mutation was present at a frequency of 71% in low grade glioma and was identified as an independent marker for improved OS in a multivariate analysis, which confirms the previous findings in low grade glioma studies. -

Supplemental Materials ZNF281 Enhances Cardiac Reprogramming
Supplemental Materials ZNF281 enhances cardiac reprogramming by modulating cardiac and inflammatory gene expression Huanyu Zhou, Maria Gabriela Morales, Hisayuki Hashimoto, Matthew E. Dickson, Kunhua Song, Wenduo Ye, Min S. Kim, Hanspeter Niederstrasser, Zhaoning Wang, Beibei Chen, Bruce A. Posner, Rhonda Bassel-Duby and Eric N. Olson Supplemental Table 1; related to Figure 1. Supplemental Table 2; related to Figure 1. Supplemental Table 3; related to the “quantitative mRNA measurement” in Materials and Methods section. Supplemental Table 4; related to the “ChIP-seq, gene ontology and pathway analysis” and “RNA-seq” and gene ontology analysis” in Materials and Methods section. Supplemental Figure S1; related to Figure 1. Supplemental Figure S2; related to Figure 2. Supplemental Figure S3; related to Figure 3. Supplemental Figure S4; related to Figure 4. Supplemental Figure S5; related to Figure 6. Supplemental Table S1. Genes included in human retroviral ORF cDNA library. Gene Gene Gene Gene Gene Gene Gene Gene Symbol Symbol Symbol Symbol Symbol Symbol Symbol Symbol AATF BMP8A CEBPE CTNNB1 ESR2 GDF3 HOXA5 IL17D ADIPOQ BRPF1 CEBPG CUX1 ESRRA GDF6 HOXA6 IL17F ADNP BRPF3 CERS1 CX3CL1 ETS1 GIN1 HOXA7 IL18 AEBP1 BUD31 CERS2 CXCL10 ETS2 GLIS3 HOXB1 IL19 AFF4 C17ORF77 CERS4 CXCL11 ETV3 GMEB1 HOXB13 IL1A AHR C1QTNF4 CFL2 CXCL12 ETV7 GPBP1 HOXB5 IL1B AIMP1 C21ORF66 CHIA CXCL13 FAM3B GPER HOXB6 IL1F3 ALS2CR8 CBFA2T2 CIR1 CXCL14 FAM3D GPI HOXB7 IL1F5 ALX1 CBFA2T3 CITED1 CXCL16 FASLG GREM1 HOXB9 IL1F6 ARGFX CBFB CITED2 CXCL3 FBLN1 GREM2 HOXC4 IL1F7 -

Id4: an Inhibitory Function in the Control
Id4: an inhibitory function in the control of hair cell formation? Sara Johanna Margarete Weber Thesis submitted to the University College London (UCL) for the degree of Master of Philosophy The work presented in this thesis was conducted at the UCL Ear Institute between September 23rd 2013 and October 19th 2015. 1st Supervisor: Dr Nicolas Daudet UCL Ear Institute, London, UK 2nd Supervisor: Dr Stephen Price UCL Department of Cell and Developmental Biology, London, UK 3rd Supervisor: Prof Guy Richardson School of Life Sciences, University of Sussex, Brighton, UK I, Sara Weber confirm that the work presented in this thesis is my own. Where information has been derived from other sources, I confirm that this has been indicated in the thesis. Heidelberg, 08.03.2016 ………………………….. Sara Weber 2 Abstract Mechanosensitive hair cells in the sensory epithelia of the vertebrate inner ear are essential for hearing and the sense of balance. Initially formed during embryological development they are constantly replaced in the adult avian inner ear after hair cell damage and loss, while practically no spontaneous regeneration occurs in mammals. The detailed molecular mechanisms that regulate hair cell formation remain elusive despite the identification of a number of signalling pathways and transcription factors involved in this process. In this study I investigated the role of Inhibitor of differentiation 4 (Id4), a member of the inhibitory class V of bHLH transcription factors, in hair cell formation. I found that Id4 is expressed in both hair cells and supporting cells of the chicken and the mouse inner ear at stages that are crucial for hair cell formation.