Transcription Factors
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Genetic Screening Identifies a Component of the SWI/SNF Complex, Arid1b As a Senescence Regulator
A genetic screening identifies a component of the SWI/SNF complex, Arid1b as a senescence regulator Sadaf Khan A thesis submitted to Imperial College London for the degree of Doctor in Philosophy MRC Clinical Sciences Centre Imperial College London, School of Medicine July 2013 Statement of originality All experiments included in this thesis were performed by myself unless otherwise stated. Copyright Declaration The copyright of this thesis rests with the author and is made available under a Creative Commons Attribution Non-Commercial No Derivatives license. Researchers are free to copy, distribute or transmit the thesis on the condition that they attribute it, that they do not use it for commercial purposes and that they do not alter, transform or build upon it. For any reuse or redistribution, researchers must make clear to others the license terms of this work. 2 Abstract Senescence is an important tumour suppressor mechanism, which prevents the proliferation of stressed or damaged cells. The use of RNA interference to identify genes with a role in senescence is an important tool in the discovery of novel cancer genes. In this work, a protocol was established for conducting bypass of senescence screenings, using shRNA libraries together with next-generation sequencing. Using this approach, the SWI/SNF subunit Arid1b was identified as a regulator of cellular lifespan in MEFs. SWI/SNF is a large multi-subunit complex that remodels chromatin. Mutations in SWI/SNF proteins are frequently associated with cancer, suggesting that SWI/SNF components are tumour suppressors. Here the role of ARID1B during senescence was investigated. Depletion of ARID1B extends the proliferative capacity of primary mouse and human fibroblasts. -

Supplementary Materials: Evaluation of Cytotoxicity and Α-Glucosidase Inhibitory Activity of Amide and Polyamino-Derivatives of Lupane Triterpenoids
Supplementary Materials: Evaluation of cytotoxicity and α-glucosidase inhibitory activity of amide and polyamino-derivatives of lupane triterpenoids Oxana B. Kazakova1*, Gul'nara V. Giniyatullina1, Akhat G. Mustafin1, Denis A. Babkov2, Elena V. Sokolova2, Alexander A. Spasov2* 1Ufa Institute of Chemistry of the Ufa Federal Research Centre of the Russian Academy of Sciences, 71, pr. Oktyabrya, 450054 Ufa, Russian Federation 2Scientific Center for Innovative Drugs, Volgograd State Medical University, Novorossiyskaya st. 39, Volgograd 400087, Russian Federation Correspondence Prof. Dr. Oxana B. Kazakova Ufa Institute of Chemistry of the Ufa Federal Research Centre of the Russian Academy of Sciences 71 Prospeсt Oktyabrya Ufa, 450054 Russian Federation E-mail: [email protected] Prof. Dr. Alexander A. Spasov Scientific Center for Innovative Drugs of the Volgograd State Medical University 39 Novorossiyskaya st. Volgograd, 400087 Russian Federation E-mail: [email protected] Figure S1. 1H and 13C of compound 2. H NH N H O H O H 2 2 Figure S2. 1H and 13C of compound 4. NH2 O H O H CH3 O O H H3C O H 4 3 Figure S3. Anticancer screening data of compound 2 at single dose assay 4 Figure S4. Anticancer screening data of compound 7 at single dose assay 5 Figure S5. Anticancer screening data of compound 8 at single dose assay 6 Figure S6. Anticancer screening data of compound 9 at single dose assay 7 Figure S7. Anticancer screening data of compound 12 at single dose assay 8 Figure S8. Anticancer screening data of compound 13 at single dose assay 9 Figure S9. Anticancer screening data of compound 14 at single dose assay 10 Figure S10. -
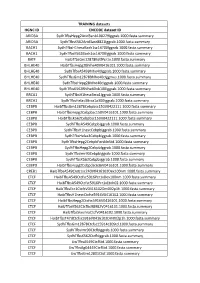
TRAINING Datasets HGNC ID ENCODE Dataset ID ARID3A
TRAINING datasets HGNC ID ENCODE dataset ID ARID3A SydhT+sHepg2Arid3anb100279Iggrab.1000.fasta.summary ARID3A SydhT+sK562Arid3asC8821Iggrab.1000.fasta.summary BACH1 SydhT+sH1hesCBaCh1sC14700Iggrab.1000.fasta.summary BACH1 SydhT+sK562BaCh1sC14700Iggrab.1000.fasta.summary BATF HaibT+sGm12878BaJPCr1x.1000.fasta.summary BHLHE40 HaibT+sHepg2Bhlhe40V0416101.1000.fasta.summary BHLHE40 SydhT+sA549Bhlhe40Iggrab.1000.fasta.summary BHLHE40 SydhT+sGm12878Bhlhe40CIggmus.1000.fasta.summary BHLHE40 SydhT+sHepg2Bhlhe40CIggrab.1000.fasta.summary BHLHE40 SydhT+sK562Bhlhe40nb100Iggrab.1000.fasta.summary BRCA1 SydhT+sH1hesCBrCa1Iggrab.1000.fasta.summary BRCA1 SydhT+sHelas3BrCa1a300Iggrab.1000.fasta.summary CEBPB HaibT+sGm12878CebpbsC150V0422111.1000.fasta.summary CEBPB HaibT+sHepg2CebpbsC150V0416101.1000.fasta.summary CEBPB HaibT+sK562CebpbsC150V0422111.1000.fasta.summary CEBPB SydhT+sA549CebpbIggrab.1000.fasta.summary CEBPB SydhT+sH1hesCCebpbIggrab.1000.fasta.summary CEBPB SydhT+sHelas3CebpbIggrab.1000.fasta.summary CEBPB SydhT+sHepg2CebpbForsklnStd.1000.fasta.summary CEBPB SydhT+sHepg2CebpbIggrab.1000.fasta.summary CEBPB SydhT+sImr90CebpbIggrab.1000.fasta.summary CEBPB SydhT+sK562CebpbIggrab.1000.fasta.summary CEBPD HaibT+sHepg2CebpdsC636V0416101.1000.fasta.summary CREB1 HaibT+sA549Creb1sC240V0416102Dex100nm.1000.fasta.summary CTCF HaibT+sA549CtCfsC5916PCr1xDex100nm.1000.fasta.summary CTCF HaibT+sA549CtCfsC5916PCr1xEtoh02.1000.fasta.summary CTCF HaibT+sECC1CtCfCV0416102Dm002p1h.1000.fasta.summary CTCF HaibT+sH1hesCCtCfsC5916V0416102.1000.fasta.summary -

1 the Neuroprotective Transcription Factor ATF5 Is Decreased And
The neuroprotective transcription factor ATF5 is decreased and sequestered into polyglutamine inclusions in Huntington’s disease Ivó H. Hernández1,2,3, Jesús Torres-Peraza1,2,5, María Santos-Galindo1,2, Eloísa Ramos-Morón4, María R. Fernández-Fernández1,2, María J. Pérez-Álvarez1,2,3, Antonio Miranda-Vizuete4 and José J. Lucas1,2* 1 Centro de Biología Molecular Severo Ochoa (CBM”SO”) CSIC/UAM, Madrid, Spain. 2 Instituto de Salud Carlos III, Networking Research Center on Neurodegenerative Diseases (CIBERNED), Spain. 3 Departamento de Biología, Facultad de Ciencias, Universidad Autónoma de Madrid, Madrid, Spain. 4 Instituto de Biomedicina de Sevilla, Hospital Universitario Virgen del Rocío/CSIC/Universidad de Sevilla, 41013 Sevilla, Spain 5 Present Address: Gerència d’Atenció Primària del Servei de Salut de les Illes Balears (IB-SALUT), Palma, Spain *Corresponding author: José J. Lucas [email protected] http://www.cbm.uam.es/lineas/lucasgroup.htm provided by Digital.CSIC View metadata, citation and similar papers at core.ac.uk CORE brought to you by 1 Abstract Activating transcription factor-5 (ATF5) is a stress-response transcription factor induced upon different cell stressors like fasting, amino-acid limitation, cadmium or arsenite. ATF5 is also induced, and promotes transcription of anti-apoptotic target genes like MCL1, during the unfolded protein response (UPR) triggered by endoplasmic reticulum stress. In the brain, high ATF5 levels are found in gliomas and also in neural progenitor cells, which need to decrease their ATF5 levels for differentiation into mature neurons or glia. This initially led to believe that ATF5 is not expressed in adult neurons. More recently, we reported basal neuronal ATF5 expression in adult mouse brain and its neuroprotective induction during UPR in a mouse model of status epilepticus. -

Roles of the CSE1L-Mediated Nuclear Import Pathway in Epigenetic
Roles of the CSE1L-mediated nuclear import pathway PNAS PLUS in epigenetic silencing Qiang Donga,b,c, Xiang Lia,b,c, Cheng-Zhi Wangb, Shaohua Xuc, Gang Yuanc, Wei Shaoc, Baodong Liud, Yong Zhengb, Hailin Wangd, Xiaoguang Leic,e,f, Zhuqiang Zhangb,1, and Bing Zhua,b,g,1 aGraduate Program, Peking Union Medical College and Chinese Academy of Medical Sciences, 100730 Beijing, China; bNational Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, 100101 Beijing, China; cNational Institute of Biological Sciences, 102206 Beijing, China; dThe State Key Laboratory of Environmental Chemistry and Ecotoxicology, Research Center for Eco-Environmental Sciences, Chinese Academy of Sciences, 100085 Beijing, China; eBeijing National Laboratory for Molecular Sciences, Department of Chemical Biology, College of Chemistry and Molecular Engineering, Peking University, Beijing 100871, China; fPeking-Tsinghua Center for Life Sciences, Peking University, 100871 Beijing, China; and gCollege of Life Sciences, University of Chinese Academy of Sciences, 100049 Beijing, China Edited by Arthur D. Riggs, Beckman Research Institute of City of Hope, Duarte, CA, and approved March 21, 2018 (received for review January 17, 2018) Epigenetic silencing can be mediated by various mechanisms, CSE1L, a key player in the nuclear import pathway, as an es- and many regulators remain to be identified. Here, we report a sential factor for maintaining the repression of many methyl- genome-wide siRNA screening to identify regulators essential for ated genes. Mechanistically, CSE1L functions by facilitating maintaining gene repression of a CMV promoter silenced by DNA the nuclear import of certain cargo proteins that are essential methylation. -

Datasheet PB1029 Anti-AEBP2 Antibody
Product datasheet Anti-AEBP2 Antibody Catalog Number: PB1029 BOSTER BIOLOGICAL TECHNOLOGY Special NO.1, International Enterprise Center, 2nd Guanshan Road, Wuhan, China Web: www.boster.com.cn Phone: +86 27 67845390 Fax: +86 27 67845390 Email: [email protected] Basic Information Product Name Anti-AEBP2 Antibody Gene Name AEBP2 Source Rabbit IgG Species Reactivity human,mouse,rat Tested Application WB,IHC-P,ICC/IF,FCM Contents 500ug/ml antibody with PBS ,0.02% NaN3 , 1mg BSA and 50% glycerol. Immunogen E.coli-derived human AEBP2 recombinant protein (Position: K424-Q517). Human AEBP2 shares 98.8% amino acid (aa) sequence identity with mouse AEBP2. Purification Immunogen affinity purified. Observed MW 54KD Dilution Ratios Western blot: 1:500-2000 Immunohistochemistry(Paraffin-embedded Section): 1:50-400 Immunocytochemistry/Immunofluorescence (ICC/IF): 1:50-400 Flow cytometry (FCM): 1-3μg/1x106 cells Storage 12 months from date of receipt,-20℃ as supplied.6 months 2 to 8℃ after reconstitution. Avoid repeated freezing and thawing Background Information Adipocyte Enhancer-Binding Protein is a zinc finger protein that in humans is encoded by the evolutionarily well-conserved gene AEBP2. This gene is mapped to 12p12.3. AEBP2 is a DNA-binding transcriptional repressor. It may regulate the migration and development of the neural crest cells through the PRC2-mediated epigenetic mechanism and is most likely a targeting protein for the mammalian PRC2 complex. Reference Anti-AEBP2 Antibody被引用在0文献中。 暂无引用 FOR RESEARCH USE ONLY. NOT FOR DIAGNOSTIC AND CLINICAL USE. 1 Product datasheet Anti-AEBP2 Antibody Catalog Number: PB1029 BOSTER BIOLOGICAL TECHNOLOGY Special NO.1, International Enterprise Center, 2nd Guanshan Road, Wuhan, China Web: www.boster.com.cn Phone: +86 27 67845390 Fax: +86 27 67845390 Email: [email protected] Selected Validation Data Figure 1. -

Examination of the Transcription Factors Acting in Bone Marrow
THESIS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY (PHD) Examination of the transcription factors acting in bone marrow derived macrophages by Gergely Nagy Supervisor: Dr. Endre Barta UNIVERSITY OF DEBRECEN DOCTORAL SCHOOL OF MOLECULAR CELL AND IMMUNE BIOLOGY DEBRECEN, 2016 Table of contents Table of contents ........................................................................................................................ 2 1. Introduction ............................................................................................................................ 5 1.1. Transcriptional regulation ................................................................................................... 5 1.1.1. Transcriptional initiation .................................................................................................. 5 1.1.2. Co-regulators and histone modifications .......................................................................... 8 1.2. Promoter and enhancer sequences guiding transcription factors ...................................... 11 1.2.1. General transcription factors .......................................................................................... 11 1.2.2. The ETS superfamily ..................................................................................................... 17 1.2.3. The AP-1 and CREB proteins ........................................................................................ 20 1.2.4. Other promoter specific transcription factor families ................................................... -

ARID1B Is a Specific Vulnerability in ARID1A-Mutant Cancers The
ARID1B is a specific vulnerability in ARID1A-mutant cancers The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters. Citation Helming, K. C., X. Wang, B. G. Wilson, F. Vazquez, J. R. Haswell, H. E. Manchester, Y. Kim, et al. 2014. “ARID1B is a specific vulnerability in ARID1A-mutant cancers.” Nature medicine 20 (3): 251-254. doi:10.1038/nm.3480. http://dx.doi.org/10.1038/nm.3480. Published Version doi:10.1038/nm.3480 Accessed February 16, 2015 10:04:32 PM EST Citable Link http://nrs.harvard.edu/urn-3:HUL.InstRepos:12987227 Terms of Use This article was downloaded from Harvard University's DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http://nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA (Article begins on next page) NIH Public Access Author Manuscript Nat Med. Author manuscript; available in PMC 2014 September 01. NIH-PA Author ManuscriptPublished NIH-PA Author Manuscript in final edited NIH-PA Author Manuscript form as: Nat Med. 2014 March ; 20(3): 251–254. doi:10.1038/nm.3480. ARID1B is a specific vulnerability in ARID1A-mutant cancers Katherine C. Helming1,2,3,4,*, Xiaofeng Wang1,2,3,*, Boris G. Wilson1,2,3, Francisca Vazquez5, Jeffrey R. Haswell1,2,3, Haley E. Manchester1,2,3, Youngha Kim1,2,3, Gregory V. Kryukov5, Mahmoud Ghandi5, Andrew J. Aguirre5,6,7, Zainab Jagani8, Zhong Wang9, Levi A. Garraway6, William C. Hahn6,7, and Charles W. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

A Clinicopathological and Molecular Genetic Analysis of Low-Grade Glioma in Adults
A CLINICOPATHOLOGICAL AND MOLECULAR GENETIC ANALYSIS OF LOW-GRADE GLIOMA IN ADULTS Presented by ANUSHREE SINGH MSc A thesis submitted in partial fulfilment of the requirements of the University of Wolverhampton for the degree of Doctor of Philosophy Brain Tumour Research Centre Research Institute in Healthcare Sciences Faculty of Science and Engineering University of Wolverhampton November 2014 i DECLARATION This work or any part thereof has not previously been presented in any form to the University or to any other body whether for the purposes of assessment, publication or for any other purpose (unless otherwise indicated). Save for any express acknowledgments, references and/or bibliographies cited in the work, I confirm that the intellectual content of the work is the result of my own efforts and of no other person. The right of Anushree Singh to be identified as author of this work is asserted in accordance with ss.77 and 78 of the Copyright, Designs and Patents Act 1988. At this date copyright is owned by the author. Signature: Anushree Date: 30th November 2014 ii ABSTRACT The aim of the study was to identify molecular markers that can determine progression of low grade glioma. This was done using various approaches such as IDH1 and IDH2 mutation analysis, MGMT methylation analysis, copy number analysis using array comparative genomic hybridisation and identification of differentially expressed miRNAs using miRNA microarray analysis. IDH1 mutation was present at a frequency of 71% in low grade glioma and was identified as an independent marker for improved OS in a multivariate analysis, which confirms the previous findings in low grade glioma studies. -

Figure S1. Representative Report Generated by the Ion Torrent System Server for Each of the KCC71 Panel Analysis and Pcafusion Analysis
Figure S1. Representative report generated by the Ion Torrent system server for each of the KCC71 panel analysis and PCaFusion analysis. (A) Details of the run summary report followed by the alignment summary report for the KCC71 panel analysis sequencing. (B) Details of the run summary report for the PCaFusion panel analysis. A Figure S1. Continued. Representative report generated by the Ion Torrent system server for each of the KCC71 panel analysis and PCaFusion analysis. (A) Details of the run summary report followed by the alignment summary report for the KCC71 panel analysis sequencing. (B) Details of the run summary report for the PCaFusion panel analysis. B Figure S2. Comparative analysis of the variant frequency found by the KCC71 panel and calculated from publicly available cBioPortal datasets. For each of the 71 genes in the KCC71 panel, the frequency of variants was calculated as the variant number found in the examined cases. Datasets marked with different colors and sample numbers of prostate cancer are presented in the upper right. *Significantly high in the present study. Figure S3. Seven subnetworks extracted from each of seven public prostate cancer gene networks in TCNG (Table SVI). Blue dots represent genes that include initial seed genes (parent nodes), and parent‑child and child‑grandchild genes in the network. Graphical representation of node‑to‑node associations and subnetwork structures that differed among and were unique to each of the seven subnetworks. TCNG, The Cancer Network Galaxy. Figure S4. REVIGO tree map showing the predicted biological processes of prostate cancer in the Japanese. Each rectangle represents a biological function in terms of a Gene Ontology (GO) term, with the size adjusted to represent the P‑value of the GO term in the underlying GO term database. -

Supplemental Materials ZNF281 Enhances Cardiac Reprogramming
Supplemental Materials ZNF281 enhances cardiac reprogramming by modulating cardiac and inflammatory gene expression Huanyu Zhou, Maria Gabriela Morales, Hisayuki Hashimoto, Matthew E. Dickson, Kunhua Song, Wenduo Ye, Min S. Kim, Hanspeter Niederstrasser, Zhaoning Wang, Beibei Chen, Bruce A. Posner, Rhonda Bassel-Duby and Eric N. Olson Supplemental Table 1; related to Figure 1. Supplemental Table 2; related to Figure 1. Supplemental Table 3; related to the “quantitative mRNA measurement” in Materials and Methods section. Supplemental Table 4; related to the “ChIP-seq, gene ontology and pathway analysis” and “RNA-seq” and gene ontology analysis” in Materials and Methods section. Supplemental Figure S1; related to Figure 1. Supplemental Figure S2; related to Figure 2. Supplemental Figure S3; related to Figure 3. Supplemental Figure S4; related to Figure 4. Supplemental Figure S5; related to Figure 6. Supplemental Table S1. Genes included in human retroviral ORF cDNA library. Gene Gene Gene Gene Gene Gene Gene Gene Symbol Symbol Symbol Symbol Symbol Symbol Symbol Symbol AATF BMP8A CEBPE CTNNB1 ESR2 GDF3 HOXA5 IL17D ADIPOQ BRPF1 CEBPG CUX1 ESRRA GDF6 HOXA6 IL17F ADNP BRPF3 CERS1 CX3CL1 ETS1 GIN1 HOXA7 IL18 AEBP1 BUD31 CERS2 CXCL10 ETS2 GLIS3 HOXB1 IL19 AFF4 C17ORF77 CERS4 CXCL11 ETV3 GMEB1 HOXB13 IL1A AHR C1QTNF4 CFL2 CXCL12 ETV7 GPBP1 HOXB5 IL1B AIMP1 C21ORF66 CHIA CXCL13 FAM3B GPER HOXB6 IL1F3 ALS2CR8 CBFA2T2 CIR1 CXCL14 FAM3D GPI HOXB7 IL1F5 ALX1 CBFA2T3 CITED1 CXCL16 FASLG GREM1 HOXB9 IL1F6 ARGFX CBFB CITED2 CXCL3 FBLN1 GREM2 HOXC4 IL1F7