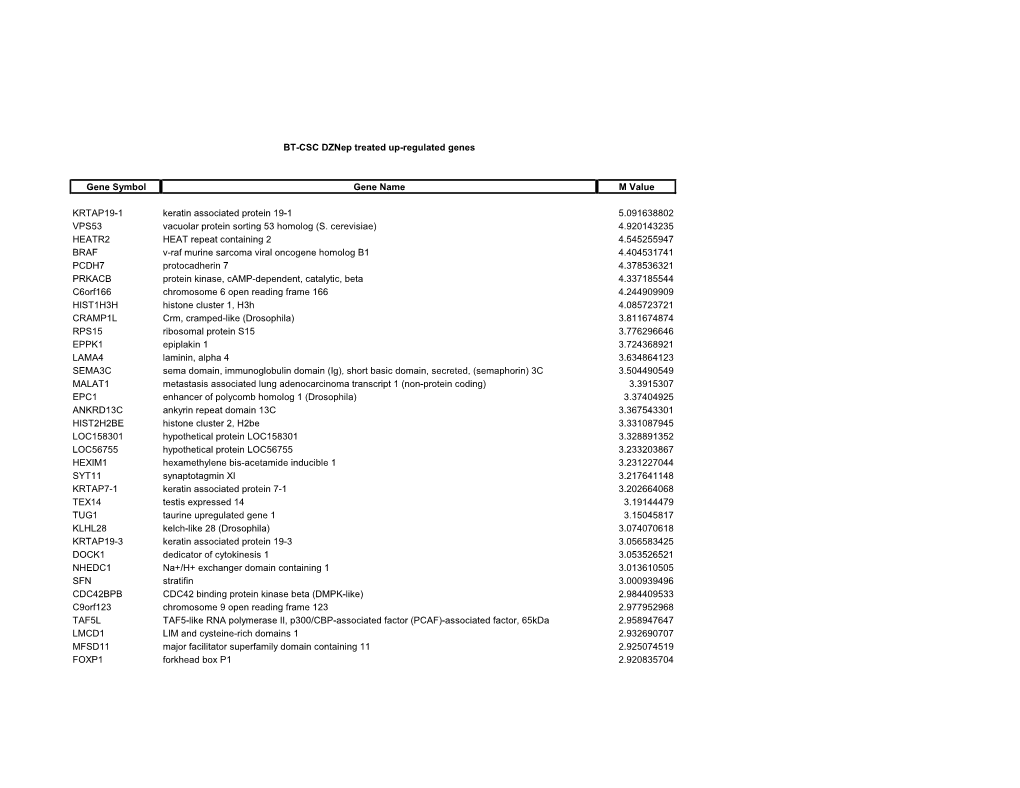
BT-CSC Dznep Treated Up-Regulated Genes Gene Symbol Gene Name M
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Plakins, a Versatile Family of Cytolinkers: Roles in Skin Integrity and in Human Diseases Jamal-Eddine Bouameur1,2, Bertrand Favre1 and Luca Borradori1
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector REVIEW Plakins, a Versatile Family of Cytolinkers: Roles in Skin Integrity and in Human Diseases Jamal-Eddine Bouameur1,2, Bertrand Favre1 and Luca Borradori1 The plakin family consists of giant proteins involved in in (Roper et al., 2002; Jefferson et al., 2004; Sonnenberg and the cross-linking and organization of the cytoskeleton Liem, 2007; Boyer et al., 2010; Suozzi et al., 2012). and adhesion complexes. They further modulate sev- Mammalian plakins share a similar structural organization eral fundamental biological processes, such as cell and comprise seven members: bullous pemphigoid antigen 1 adhesion, migration, and polarization or signaling (BPAG1), desmoplakin, envoplakin, epiplakin, microtubule- pathways. Inherited and acquired defects of plakins actin cross-linking factor 1 (MACF1), periplakin, and plectin in humans and in animal models potentially lead to (Figure 1) (Choi et al., 2002; Jefferson et al., 2007; Choi and dramatic manifestations in the skin, striated muscles, Weis, 2011; Ortega et al., 2011). The existence of develop- and/or nervous system. These observations unequivo- mentally regulated and tissue-specific splice variants of some cally demonstrate the key role of plakins in the plakins further increases the diversity and versatility of these proteins (Table 1; Figure 1; Leung et al., 2001; Rezniczek et al., maintenance of tissue integrity. Here we review the 2003; Lin et al., 2005; Jefferson et al., 2006; Cabral et al., 2010). characteristics of the mammalian plakin members BPAG1 (bullous pemphigoid antigen 1), desmoplakin, PLAKINS IN THE EPIDERMIS plectin, envoplakin, epiplakin, MACF1 (microtubule- Epithelial BPAG1 (BPAG1e, also called BP230) constitutes the actin cross-linking factor 1), and periplakin, highlight- epithelium-specific isoform of BPAG1 and is localized in basal ing their role in skin homeostasis and diseases. -

How Shelterin Protects Mammalian Telomeres 303 ANRV361-GE42-15 ARI 3 October 2008 10:10 (See 3' 5' TRF2 S Mplex
ANRV361-GE42-15 ARI 3 October 2008 10:10 ANNUAL How Shelterin Protects REVIEWS Further Click here for quick links to Annual Reviews content online, Mammalian Telomeres including: • Other articles in this volume 1 • Top cited articles Wilhelm Palm and Titia de Lange • Top downloaded articles • Our comprehensive search Laboratory for Cell Biology and Genetics, The Rockefeller University, New York, NY 10021; email: [email protected] 1Current address: Max Planck Institute of Molecular Cell Biology and Genetics, 01307 Dresden, Germany Annu. Rev. Genet. 2008. 42:301–34 Key Words First published online as a Review in Advance on ATM, ATR, cancer, NHEJ, HR August 4, 2008 by Rockefeller University on 10/13/09. For personal use only. The Annual Review of Genetics is online at Abstract genet.annualreviews.org The genomes of prokaryotes and eukaryotic organelles are usually cir- This article’s doi: cular as are most plasmids and viral genomes. In contrast, the nuclear Annu. Rev. Genet. 2008.42:301-334. Downloaded from arjournals.annualreviews.org 10.1146/annurev.genet.41.110306.130350 genomes of eukaryotes are organized on linear chromosomes, which Copyright c 2008 by Annual Reviews. require mechanisms to protect and replicate DNA ends. Eukaryotes All rights reserved navigate these problemswith the advent of telomeres, protective nucle- 0066-4197/08/1201-0301$20.00 oprotein complexes at the ends of linear chromosomes, and telomerase, the enzyme that maintains the DNA in these structures. Mammalian telomeres contain a specific protein complex, shelterin, that functions to protect chromosome ends from all aspects of the DNA damage re- sponse and regulates telomere maintenance by telomerase. -

Allele-Specific Expression of Ribosomal Protein Genes in Interspecific Hybrid Catfish
Allele-specific Expression of Ribosomal Protein Genes in Interspecific Hybrid Catfish by Ailu Chen A dissertation submitted to the Graduate Faculty of Auburn University in partial fulfillment of the requirements for the Degree of Doctor of Philosophy Auburn, Alabama August 1, 2015 Keywords: catfish, interspecific hybrids, allele-specific expression, ribosomal protein Copyright 2015 by Ailu Chen Approved by Zhanjiang Liu, Chair, Professor, School of Fisheries, Aquaculture and Aquatic Sciences Nannan Liu, Professor, Entomology and Plant Pathology Eric Peatman, Associate Professor, School of Fisheries, Aquaculture and Aquatic Sciences Aaron M. Rashotte, Associate Professor, Biological Sciences Abstract Interspecific hybridization results in a vast reservoir of allelic variations, which may potentially contribute to phenotypical enhancement in the hybrids. Whether the allelic variations are related to the downstream phenotypic differences of interspecific hybrid is still an open question. The recently developed genome-wide allele-specific approaches that harness high- throughput sequencing technology allow direct quantification of allelic variations and gene expression patterns. In this work, I investigated allele-specific expression (ASE) pattern using RNA-Seq datasets generated from interspecific catfish hybrids. The objective of the study is to determine the ASE genes and pathways in which they are involved. Specifically, my study investigated ASE-SNPs, ASE-genes, parent-of-origins of ASE allele and how ASE would possibly contribute to heterosis. My data showed that ASE was operating in the interspecific catfish system. Of the 66,251 and 177,841 SNPs identified from the datasets of the liver and gill, 5,420 (8.2%) and 13,390 (7.5%) SNPs were identified as significant ASE-SNPs, respectively. -

Supplemental Table S1
Entrez Gene Symbol Gene Name Affymetrix EST Glomchip SAGE Stanford Literature HPA confirmed Gene ID Profiling profiling Profiling Profiling array profiling confirmed 1 2 A2M alpha-2-macroglobulin 0 0 0 1 0 2 10347 ABCA7 ATP-binding cassette, sub-family A (ABC1), member 7 1 0 0 0 0 3 10350 ABCA9 ATP-binding cassette, sub-family A (ABC1), member 9 1 0 0 0 0 4 10057 ABCC5 ATP-binding cassette, sub-family C (CFTR/MRP), member 5 1 0 0 0 0 5 10060 ABCC9 ATP-binding cassette, sub-family C (CFTR/MRP), member 9 1 0 0 0 0 6 79575 ABHD8 abhydrolase domain containing 8 1 0 0 0 0 7 51225 ABI3 ABI gene family, member 3 1 0 1 0 0 8 29 ABR active BCR-related gene 1 0 0 0 0 9 25841 ABTB2 ankyrin repeat and BTB (POZ) domain containing 2 1 0 1 0 0 10 30 ACAA1 acetyl-Coenzyme A acyltransferase 1 (peroxisomal 3-oxoacyl-Coenzyme A thiol 0 1 0 0 0 11 43 ACHE acetylcholinesterase (Yt blood group) 1 0 0 0 0 12 58 ACTA1 actin, alpha 1, skeletal muscle 0 1 0 0 0 13 60 ACTB actin, beta 01000 1 14 71 ACTG1 actin, gamma 1 0 1 0 0 0 15 81 ACTN4 actinin, alpha 4 0 0 1 1 1 10700177 16 10096 ACTR3 ARP3 actin-related protein 3 homolog (yeast) 0 1 0 0 0 17 94 ACVRL1 activin A receptor type II-like 1 1 0 1 0 0 18 8038 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 1 0 0 0 0 19 8751 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 1 0 0 0 0 20 8728 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 1 0 0 0 0 21 81792 ADAMTS12 ADAM metallopeptidase with thrombospondin type 1 motif, 12 1 0 0 0 0 22 9507 ADAMTS4 ADAM metallopeptidase with thrombospondin type 1 -

ANKRD11 Gene Ankyrin Repeat Domain 11
ANKRD11 gene ankyrin repeat domain 11 Normal Function The ANKRD11 gene provides instructions for making a protein called ankyrin repeat domain 11 (ANKRD11). As its name suggests, this protein contains multiple regions called ankyrin domains; proteins with these domains help other proteins interact with each other. The ANKRD11 protein interacts with certain proteins called histone deacetylases, which are important for controlling gene activity. Through these interactions, ANKRD11 affects when genes are turned on and off. For example, ANKRD11 brings together histone deacetylases and other proteins called p160 coactivators. This association regulates the ability of p160 coactivators to turn on gene activity. ANKRD11 may also enhance the activity of a protein called p53, which controls the growth and division (proliferation) and the self-destruction (apoptosis) of cells. The ANKRD11 protein is found in nerve cells (neurons) in the brain. During embryonic development, ANKRD11 helps regulate the proliferation of these cells and development of the brain. Researchers speculate that the protein may also be involved in the ability of neurons to change and adapt over time (plasticity), which is important for learning and memory. ANKRD11 may function in other cells in the body and appears to be involved in normal bone development. Health Conditions Related to Genetic Changes KBG syndrome Several ANKRD11 gene mutations have been found to cause KBG syndrome, a condition characterized by large upper front teeth and other unusual facial features, skeletal abnormalities, and intellectual disability. Most of these mutations lead to an abnormally short ANKRD11 protein, which likely has little or no function. Reduction of this protein's function is thought to underlie the signs and symptoms of the condition. -

Genetic Basis of Simple and Complex Traits with Relevance to Avian Evolution
Genetic basis of simple and complex traits with relevance to avian evolution Małgorzata Anna Gazda Doctoral Program in Biodiversity, Genetics and Evolution D Faculdade de Ciências da Universidade do Porto 2019 Supervisor Miguel Jorge Pinto Carneiro, Auxiliary Researcher, CIBIO/InBIO, Laboratório Associado, Universidade do Porto Co-supervisor Ricardo Lopes, CIBIO/InBIO Leif Andersson, Uppsala University FCUP Genetic basis of avian traits Nota Previa Na elaboração desta tese, e nos termos do número 2 do Artigo 4º do Regulamento Geral dos Terceiros Ciclos de Estudos da Universidade do Porto e do Artigo 31º do D.L.74/2006, de 24 de Março, com a nova redação introduzida pelo D.L. 230/2009, de 14 de Setembro, foi efetuado o aproveitamento total de um conjunto coerente de trabalhos de investigação já publicados ou submetidos para publicação em revistas internacionais indexadas e com arbitragem científica, os quais integram alguns dos capítulos da presente tese. Tendo em conta que os referidos trabalhos foram realizados com a colaboração de outros autores, o candidato esclarece que, em todos eles, participou ativamente na sua conceção, na obtenção, análise e discussão de resultados, bem como na elaboração da sua forma publicada. Este trabalho foi apoiado pela Fundação para a Ciência e Tecnologia (FCT) através da atribuição de uma bolsa de doutoramento (PD/BD/114042/2015) no âmbito do programa doutoral em Biodiversidade, Genética e Evolução (BIODIV). 2 FCUP Genetic basis of avian traits Acknowledgements Firstly, I would like to thank to my all supervisors Miguel Carneiro, Ricardo Lopes and Leif Andersson, for the demanding task of supervising myself last four years. -

Autism Multiplex Family with 16P11.2P12.2 Microduplication Syndrome in Monozygotic Twins and Distal 16P11.2 Deletion in Their Brother
European Journal of Human Genetics (2012) 20, 540–546 & 2012 Macmillan Publishers Limited All rights reserved 1018-4813/12 www.nature.com/ejhg ARTICLE Autism multiplex family with 16p11.2p12.2 microduplication syndrome in monozygotic twins and distal 16p11.2 deletion in their brother Anne-Claude Tabet1,2,3,4, Marion Pilorge2,3,4, Richard Delorme5,6,Fre´de´rique Amsellem5,6, Jean-Marc Pinard7, Marion Leboyer6,8,9, Alain Verloes10, Brigitte Benzacken1,11,12 and Catalina Betancur*,2,3,4 The pericentromeric region of chromosome 16p is rich in segmental duplications that predispose to rearrangements through non-allelic homologous recombination. Several recurrent copy number variations have been described recently in chromosome 16p. 16p11.2 rearrangements (29.5–30.1 Mb) are associated with autism, intellectual disability (ID) and other neurodevelopmental disorders. Another recognizable but less common microdeletion syndrome in 16p11.2p12.2 (21.4 to 28.5–30.1 Mb) has been described in six individuals with ID, whereas apparently reciprocal duplications, studied by standard cytogenetic and fluorescence in situ hybridization techniques, have been reported in three patients with autism spectrum disorders. Here, we report a multiplex family with three boys affected with autism, including two monozygotic twins carrying a de novo 16p11.2p12.2 duplication of 8.95 Mb (21.28–30.23 Mb) characterized by single-nucleotide polymorphism array, encompassing both the 16p11.2 and 16p11.2p12.2 regions. The twins exhibited autism, severe ID, and dysmorphic features, including a triangular face, deep-set eyes, large and prominent nasal bridge, and tall, slender build. The eldest brother presented with autism, mild ID, early-onset obesity and normal craniofacial features, and carried a smaller, overlapping 16p11.2 microdeletion of 847 kb (28.40–29.25 Mb), inherited from his apparently healthy father. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Gene Expression Imputation Across Multiple Brain Regions Provides Insights Into Schizophrenia Risk
VU Research Portal Gene expression imputation across multiple brain regions provides insights into schizophrenia risk iPSYCH-GEMS Schizophrenia Working Group; CommonMind Consortium; The Schizophrenia Working Group of the PsyUniversity of Copenhagenchiatric Genomics Consortium published in Nature Genetics 2019 DOI (link to publisher) 10.1038/s41588-019-0364-4 document version Publisher's PDF, also known as Version of record document license Article 25fa Dutch Copyright Act Link to publication in VU Research Portal citation for published version (APA) iPSYCH-GEMS Schizophrenia Working Group, CommonMind Consortium, & The Schizophrenia Working Group of the PsyUniversity of Copenhagenchiatric Genomics Consortium (2019). Gene expression imputation across multiple brain regions provides insights into schizophrenia risk. Nature Genetics, 51(4), 659–674. https://doi.org/10.1038/s41588-019-0364-4 General rights Copyright and moral rights for the publications made accessible in the public portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. • Users may download and print one copy of any publication from the public portal for the purpose of private study or research. • You may not further distribute the material or use it for any profit-making activity or commercial gain • You may freely distribute the URL identifying the publication in the public portal ? Take down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. E-mail address: [email protected] Download date: 28. -

Epigenetic Regulation of the Human Genome by Transposable Elements
EPIGENETIC REGULATION OF THE HUMAN GENOME BY TRANSPOSABLE ELEMENTS A Dissertation Presented to The Academic Faculty By Ahsan Huda In Partial Fulfillment Of the Requirements for the Degree Doctor of Philosophy in Bioinformatics in the School of Biology Georgia Institute of Technology August 2010 EPIGENETIC REGULATION OF THE HUMAN GENOME BY TRANSPOSABLE ELEMENTS Approved by: Dr. I. King Jordan, Advisor Dr. John F. McDonald School of Biology School of Biology Georgia Institute of Technology Georgia Institute of Technology Dr. Leonardo Mariño-Ramírez Dr. Jung Choi NCBI/NLM/NIH School of Biology Georgia Institute of Technology Dr. Soojin Yi, School of Biology Georgia Institute of Technology Date Approved: June 25, 2010 To my mother, your life is my inspiration... ACKNOWLEDGEMENTS I am evermore thankful to my advisor Dr. I. King Jordan for his guidance, support and encouragement throughout my years as a PhD student. I am very fortunate to have him as my mentor as he is instrumental in shaping my personal and professional development. His contributions will continue to impact my life and career and for that I am forever grateful. I am also thankful to my committee members, John McDonald, Leonardo Mariño- Ramírez, Soojin Yi and Jung Choi for their continued support during my PhD career. Through my meetings and discussions with them, I have developed an appreciation for the scientific method and a thorough understanding of my field of study. I am especially grateful to my friends and colleagues, Lee Katz and Jittima Piriyapongsa for their support and presence, which brightened the atmosphere in the lab in the months and years past. -

Ingenuity Pathway Analysis of Differentially Expressed Genes Involved in Signaling Pathways and Molecular Networks in Rhoe Gene‑Edited Cardiomyocytes
INTERNATIONAL JOURNAL OF MOleCular meDICine 46: 1225-1238, 2020 Ingenuity pathway analysis of differentially expressed genes involved in signaling pathways and molecular networks in RhoE gene‑edited cardiomyocytes ZHONGMING SHAO1*, KEKE WANG1*, SHUYA ZHANG2, JIANLING YUAN1, XIAOMING LIAO1, CAIXIA WU1, YUAN ZOU1, YANPING HA1, ZHIHUA SHEN1, JUNLI GUO2 and WEI JIE1,2 1Department of Pathology, School of Basic Medicine Sciences, Guangdong Medical University, Zhanjiang, Guangdong 524023; 2Hainan Provincial Key Laboratory for Tropical Cardiovascular Diseases Research and Key Laboratory of Emergency and Trauma of Ministry of Education, Institute of Cardiovascular Research of The First Affiliated Hospital, Hainan Medical University, Haikou, Hainan 571199, P.R. China Received January 7, 2020; Accepted May 20, 2020 DOI: 10.3892/ijmm.2020.4661 Abstract. RhoE/Rnd3 is an atypical member of the Rho super- injury and abnormalities, cell‑to‑cell signaling and interaction, family of proteins, However, the global biological function and molecular transport. In addition, 885 upstream regulators profile of this protein remains unsolved. In the present study, a were enriched, including 59 molecules that were predicated RhoE‑knockout H9C2 cardiomyocyte cell line was established to be strongly activated (Z‑score >2) and 60 molecules that using CRISPR/Cas9 technology, following which differentially were predicated to be significantly inhibited (Z‑scores <‑2). In expressed genes (DEGs) between the knockout and wild‑type particular, 33 regulatory effects and 25 networks were revealed cell lines were screened using whole genome expression gene to be associated with the DEGs. Among them, the most signifi- chips. A total of 829 DEGs, including 417 upregulated and cant regulatory effects were ‘adhesion of endothelial cells’ and 412 downregulated, were identified using the threshold of ‘recruitment of myeloid cells’ and the top network was ‘neuro- fold changes ≥1.2 and P<0.05. -

Dyskerin Mutations Present in Dyskeratosis Congenita Patients Increase Oxidative Stress and DNA Damage Signalling in Dictyostelium Discoideum
cells Article Dyskerin Mutations Present in Dyskeratosis Congenita Patients Increase Oxidative Stress and DNA Damage Signalling in Dictyostelium Discoideum Javier Rodriguez-Centeno, Rosario Perona and Leandro Sastre * Instituto de Investigaciones Biomedicas, CSIC/UAM and Centro de Investigación Biomédica en Red de Enfermedades Raras, CIBERER, 28029 Madrid, Spain; [email protected] (J.R.-C.); [email protected] (R.P.) * Correspondence: [email protected]; Tel.: +3491-585-4437 Received: 11 October 2019; Accepted: 5 November 2019; Published: 8 November 2019 Abstract: Dyskerin is a protein involved in the formation of small nucleolar and small Cajal body ribonucleoproteins. These complexes participate in RNA pseudouridylation and are also components of the telomerase complex required for telomere elongation. Dyskerin mutations cause a rare disease, X-linked dyskeratosis congenita, with no curative treatment. The social amoeba Dictyostelium discoideum contains a gene coding for a dyskerin homologous protein. In this article D. discoideum mutant strains that have mutations corresponding to mutations found in dyskeratosis congenita patients are described. The phenotype of the mutant strains has been studied and no alterations were observed in pseudouridylation activity and telomere structure. Mutant strains showed increased proliferation on liquid culture but reduced growth feeding on bacteria. The results obtained indicated the existence of increased DNA damage response and reactive oxygen species, as also reported in human Dyskeratosis congenita cells and some other disease models. These data, together with the haploid character of D. discoideum vegetative cells, that resemble the genomic structure of the human dyskerin gene, located in the X chromosome, support the conclusion that D. discoideum can be a good model system for the study of this disease.