In Vitro Measurements of Cellular Forces and Their Importance in the Lung—From the Sub- to the Multicellular Scale
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Attachment 1
Appendix 1 Chemico-Biological Interactions 301 (2019) 2–5 Contents lists available at ScienceDirect Chemico-Biological Interactions journal homepage: www.elsevier.com/locate/chembioint An examination of the linear no-threshold hypothesis of cancer risk T assessment: Introduction to a series of reviews documenting the lack of biological plausibility of LNT R. Goldena,*, J. Busb, E. Calabresec a ToxLogic, Gaithersburg, MD, USA b Exponent, Midland, MI, USA c University of Massachusetts, Amherst, MA, USA The linear no-threshold (LNT) single-hit dose response model for evolution and the prominence of co-author Gilbert Lewis, who would be mutagenicity and carcinogenicity has dominated the field of regulatory nominated for the Nobel Prize some 42 times, this idea generated much risk assessment of carcinogenic agents since 1956 for radiation [8] and heat but little light. This hypothesis was soon found to be unable to 1977 for chemicals [11]. The fundamental biological assumptions upon account for spontaneous mutation rates, underestimating such events which the LNT model relied at its early adoption at best reflected a by a factor of greater than 1000-fold [19]. primitive understanding of key biological processes controlling muta- Despite this rather inauspicious start for the LNT model, Muller tion and development of cancer. However, breakthrough advancements would rescue it from obscurity, giving it vast public health and medical contributed by modern molecular biology over the last several decades implications, even proclaiming it a scientific principle by calling it the have provided experimental tools and evidence challenging the LNT Proportionality Rule [20]. While initially conceived as a driving force model for use in risk assessment of radiation or chemicals. -

THE NAMES LIST Version 7.2 -.:: GEOCITIES.Ws
List of Names, Vers. 7.2 THE NAMES LIST Version 7.2 We apologize if any names are misspelled, misplaced or in any other way misused – we can’t know everything! Shorter versions and/or nicknames are listed as Name (Short version) Some names are listed with their meaning in English as Name=english word All firstnames marked with an * are rather old-fashioned and not common among younger people any more. The surnames marked with an ** are names of prominent persons which are actually very rare. CONTEMPORARY NAMES (Bear in mind that several countries, China to name one, list surnames/family names before firstnames/personal names!) Song Abdullah African Surnames Songo’O Achmed Tchami Ahmet Adepoju Tchango Akif Amokachi Tchoutang Ali Amunike Tinkler Arif Andem Tovey Aykut Angibeaud Bahri Ardense Bekir Babayaro Arabian Female Firstnames Bülent Baruwa Aynur Durmus Billong Ayse Dursun Dosu Aysel Erdal Ekoku Belkis Erkan Etamé Berna Erol Finidi Cigdem Fahri Foe Damla Gazi Ikpeba Demet Halifi Ipoua Derya Halil Issa Dilek Hamza Kalla Elif Hassan Kanu Elo Hüsein Khumaleo Fahrie Iskender Kipketer Fatima Ismail Lobé Fatma Ismet M’butu Fehime Khalib Mapela Gulizar Khaled Masinga Gülcan Leyla Mboma Hacer Mehmet Mimboe Hatice Yusuf Moeti Hazel Zeki Moshoeu Hediye Ziya Motaung Hüeyla Moukoko Makbule Obafemi Medine Arabian Surnames Ohenen Mürüret Al - Okechukwu Nazli Okocha Shahrzad Abedzadeh Okpara Tanzu Al Awad Olembé Al Daeya Oliseh Arabian Male Firstnames Al Dokhy Opakuru Al Dossari Abdul Redebe Al Dossary Compiled By Krikkit Gamblers Page 1 List of Names, -

Wayne County Death Index, 1934-1939 1 SURNAME FIRST
SURNAME FIRST NAME MIDDLE AGE DATE LOCATION BOX FILE Aaron Phillip W 305 4/21/1938 WD 21 16 Aarons Victoria 149 10/9/1937 EC 13 16 Abbeg Minnie Manning 160 7/13/1938 SM TP 21 8 Abbey Thomas J 180 8/7/1939 HP 30 16 Abbote Vito 165 3/3/1938 NK TP 23 7 Abbott Ada 180 3/2/1938 HP 22 3 Abbott Blanche Maude 152 5/24/1938 HP 22 7 Abbott Chester 155 10/21/1938 WD 21 20 Abbott Ira W 190 8/20/1935 WY 6 11 Abbott Melvin J 102 3/31/1936 WD 9 15 Abbott Roy 142 6/24/1937 WD 15 19 Abbott William C 184 9/28/1936 RM TP 8 14 Abell Ellen 163 2/20/1937 NK TP 17 8 Abelson James H 183 11/27/1938 NK TP 24 12 Aben Augusta 171 11/25/1937 LP 14 15 Abernathy Paul 152 4/3/1939 NK TP 28 16 Abjorson Sven J 148 8/2/1938 GPF 19 16 Abraham Anna 155 4/3/1938 HP 22 5 Abraham Fay 142 9/17/1939 WD 27 22 Abraham Frank Wilson 119 2/18/1937 HP 16 4 Abraham Omar 150 8/2/1938 NK TP 24 1 Abrahamson John 157 12/22/1937 NK TP 18 16 Abram William 175 11/3/1937 NK TP 18 12 Abshagen William 163 2/11/1937 NK TP 17 7 Acciaccafero Dora 159 5/21/1939 NK TP 28 23 Acker Baby 0 4/12/1936 ML 7 27 Acker William H 187 8/12/1939 HP 30 16 Ackerman Jessie W 165 5/31/1939 WD 27 18 Ackerman Leo 174 4/15/1935 NK TP 4 14 Ackermann Joseph M 167 4/15/1936 HP 10 7 Ackersville William 141 10/30/1935 HP 3 19 Ackley Robert D 106 6/28/1936 HP 10 12 Adach John 150 8/17/1939 NK TP 29 7 Adair Baby 0 8/28/1939 WD 27 21 Adair Kristine Ann 105 5/6/1937 WD 15 17 Adamczak Baby 0 7/8/1935 HM 1 29 Adamczak Veronika 301 1/14/1939 HM 31 1 Adamczyk Baby 0 8/3/1937 HM 14 8 Adamczyk Donald 204 2/6/1937 HM 14 2 Adamczyk -
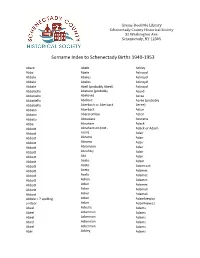
Surname Index to Schenectady Births 1940-1953
Grems-Doolittle Library Schenectady County Historical Society 32 Washington Ave. Schenectady, NY 12305 Surname Index to Schenectady Births 1940-1953 Abare Abele Ackley Abba Abele Ackroyd Abbale Abeles Ackroyd Abbale Abeles Ackroyd Abbale Abell (probably Abeel) Ackroyd Abbatiello Abelone (probably Acord Abbatiello Abelove) Acree Abbatiello Abelove Acree (probably Abbatiello Aberbach or Aberback Aeree) Abbato Aberback Acton Abbato Abercrombie Acton Abbato Aboudara Acucena Abbe Abraham Adack Abbott Abrahamson (not - Adack or Adach Abbott nson) Adair Abbott Abrams Adair Abbott Abrams Adair Abbott Abramson Adair Abbott Abrofsky Adair Abbott Abt Adair Abbott Aceto Adam Abbott Aceto Adamczak Abbott Aceto Adamec Abbott Aceto Adamec Abbott Acken Adamec Abbott Acker Adamec Abbott Acker Adamek Abbott Acker Adamek Abbzle = ? spelling Acker Adamkiewicz unclear Acker Adamkiewicz Abeel Ackerle Adams Abeel Ackerman Adams Abeel Ackerman Adams Abeel Ackerman Adams Abeel Ackerman Adams Abel Ackley Adams Grems-Doolittle Library Schenectady County Historical Society 32 Washington Ave. Schenectady, NY 12305 Surname Index to Schenectady Births 1940-1953 Adams Adamson Ahl Adams Adanti Ahles Adams Addis Ahman Adams Ademec or Adamec Ahnert Adams Adinolfi Ahren Adams Adinolfi Ahren Adams Adinolfi Ahrendtsen Adams Adinolfi Ahrendtsen Adams Adkins Ahrens Adams Adkins Ahrens Adams Adriance Ahrens Adams Adsit Aiken Adams Aeree Aiken Adams Aernecke Ailes = ? Adams Agans Ainsworth Adams Agans Aker (or Aeher = ?) Adams Aganz (Agans ?) Akers Adams Agare or Abare = ? Akerson Adams Agat Akin Adams Agat Akins Adams Agen Akins Adams Aggen Akland Adams Aggen Albanese Adams Aggen Alberding Adams Aggen Albert Adams Agnew Albert Adams Agnew Albert or Alberti Adams Agnew Alberti Adams Agostara Alberti Adams Agostara (not Agostra) Alberts Adamski Agree Albig Adamski Ahave ? = totally Albig Adamson unclear Albohm Adamson Ahern Albohm Adamson Ahl Albohm (not Albolm) Adamson Ahl Albrezzi Grems-Doolittle Library Schenectady County Historical Society 32 Washington Ave. -

The Interface Between English and the Celtic Languages
Universität Potsdam Hildegard L. C. Tristram (ed.) The Celtic Englishes IV The Interface between English and the Celtic Languages Potsdam University Press In memoriam Alan R. Thomas Contents Hildegard L.C. Tristram Inroduction .................................................................................................... 1 Alan M. Kent “Bringin’ the Dunkey Down from the Carn:” Cornu-English in Context 1549-2005 – A Provisional Analysis.................. 6 Gary German Anthroponyms as Markers of Celticity in Brittany, Cornwall and Wales................................................................. 34 Liam Mac Mathúna What’s in an Irish Name? A Study of the Personal Naming Systems of Irish and Irish English ......... 64 John M. Kirk and Jeffrey L. Kallen Irish Standard English: How Celticised? How Standardised?.................... 88 Séamus Mac Mathúna Remarks on Standardisation in Irish English, Irish and Welsh ................ 114 Kevin McCafferty Be after V-ing on the Past Grammaticalisation Path: How Far Is It after Coming? ..................................................................... 130 Ailbhe Ó Corráin On the ‘After Perfect’ in Irish and Hiberno-English................................. 152 II Contents Elvira Veselinovi How to put up with cur suas le rud and the Bidirectionality of Contact .................................................................. 173 Erich Poppe Celtic Influence on English Relative Clauses? ......................................... 191 Malcolm Williams Response to Erich Poppe’s Contribution -

Petition Signatories 12 November 2020 First Name Surname Country Capacity
Petition Signatories 12 November 2020 First name Surname Country Capacity 1 Ulisses Abade Brasil Vice Presidente Sindicato 2 Sandrine Abayou France salariée 3 HAYANI ABDEL BELGIUM trade union 4 Ariadna Abeltina Latvia Trade Union Officer General Secretary FSC 5 Roberto Abenia España CCOO Aragón 6 Pascal Abenza France Délégué Syndical Groupe 7 Jacques ADAM Luxembourg membre du syndicat 8 Lenka Adamcikova Slowakei Member of works 9 Ole Einar Adamsrød Norway Trade Union 10 Paula Adao Luxembourg Déléguée 11 Nicolae Adrian România Union member 12 Costache Adrian Alin Romania Trade union 13 Pana Adriana Laura România Member of works council 14 Bert Aerts Belgium member of works council 15 Annick Aerts Belgium trade union 16 Sascha Aerts België 200 Secrétaire Générale UL 17 Odile AGRAFEIL FRANCE CGT 18 Oscar Aguado España Miembro comité empresa 19 Fátima Aguado Queipo Spain Trade union 20 Agustin Aguila Mellado España Miembro del Sindicato SECRETARIO UGT ADIF 21 HERNANDEZ AGUILAR OSWALD BARCELONA 22 Antonio Angel Aguilar Fernández España Trade Union 23 Jaan Aiaots Estonia Trade union SYNDICAT SYNPTAC-CGT 24 Nora AINECHE FRANCE PARIS FRANCE 25 Raul Aira España miembro comité empresa 26 Alessandra Airaldi Italy TRADE union 27 Juan Miguel Aisa Spain Member of works council 28 Juan-Miguel AISA Spain EWC Membre élu du comité européen Driver Services 29 Sylvestre AISSI France Norauto 30 Sara Akervall Sweden EWC 31 Michiel Al Netherlands trade union official 32 Nickels Alain Luxemburg Trade Union 33 MAURO ALBANESE FRANCE SYNDICAT 34 Michela Albarello -

Author Index to Abstracts
Pediat. Res. 16: 365A-380A (1982) Author Index to Abstracts Numbers refer to abstract numbers Abbasi S 7 11,767,1166, 1380, 1591 Alteneder RE 97 1 Ardito T 1458 Baker H 604 Abbassi V 403 Altman AJ 7 14 Armes D 286 Baker HJ 1576 Abboud E 1606 Alvarado CS 7 15 Armstrong D 927 Baker L 236, 803, 1100, 11 12, 1133 Abman SH 1644 Alverson DC 126, 1172, 1 173, 1 174 Arnett J 1451, 1452 Baker LR 448 Abraham J 1694 Ambrosino DM 93 1, 1029 Arnold BL 30 Baker MK 1147 Abrams CK 457 Ambruso DR 661, 716 Arnold WC 477, 1434 Baldomero A 401, 103 1, 1087, 1392, Abrams N 805 Amendt BA 1 104 Arosio P 1252 1393, 1394, 1404 Abramson JS 928,964 Ament ME 466,528 Arredondo JL 935 Baldwin A I1 Accardo PJ 38, 85 Amler D 43 1 Arrobio JO 418 Baldwin C I I Accurso FJ 1644 Amma P 1019 Arroyo PJ 895 Bale JF Jr 937 Aceto T Jr 376 Amma PLS 1401, 1571 Arslanian S 33 1 Baley JE 846,938, 1437 Ackerman BA 493 Ammann AJ 847,954 Artman HG 21 1 Balfe J 1478 Ackerman M 857 Ampola MG 693 Artman M 25 1 Balfe JW 1499 Ackerman N 1298, 1667, 1703 Amsel J 1332 Arthur DC 773 Balfour HH Jr 1007 Ackerman NB Jr 1592 Amylon MD 756 Arvin AM 1048, I055 Balfour IC LO1 Ackerman R 1372, 1373 Anas NG 1593, 1594 Awin D 43 1 Balistreri WF 462, 596 Acosta PB 1145 Anday EK 59,2 12, 1299 Asinger RW 150 Ballard JL 1183 Adachihara A 760 Andersen J 1663 Aster R 729 Ballard PL 195, 1182, 1184, 1630 Adamkin DH 248,458, 1167, 1168, Andersen JM 1059, 1060 Atakent YS 432, 1203 Ballard RA 72, 1463, 1630 1169 Anderson C 1369 Aten MJ I I Ballow M 9 14 Adams JM 1387, 1388 Anderson CL 13 13 Ater JL 717 Baluarte J 21. -

Consensus Guidelines for the Use and Interpretation of Angiogenesis Assays
Angiogenesis (2018) 21:425–532 https://doi.org/10.1007/s10456-018-9613-x REVIEW PAPER Consensus guidelines for the use and interpretation of angiogenesis assays Patrycja Nowak‑Sliwinska1,2 · Kari Alitalo3 · Elizabeth Allen4 · Andrey Anisimov3 · Alfred C. Aplin5 · Robert Auerbach6 · Hellmut G. Augustin7,8,9 · David O. Bates10 · Judy R. van Beijnum11 · R. Hugh F. Bender12 · Gabriele Bergers13,4 · Andreas Bikfalvi14 · Joyce Bischof15 · Barbara C. Böck7,8,9 · Peter C. Brooks16 · Federico Bussolino17,18 · Bertan Cakir19 · Peter Carmeliet20,21 · Daniel Castranova22 · Anca M. Cimpean23 · Ondine Cleaver24 · George Coukos25 · George E. Davis26 · Michele De Palma27 · Anna Dimberg28 · Ruud P. M. Dings29 · Valentin Djonov30 · Andrew C. Dudley31,32 · Neil P. Dufton33 · Sarah‑Maria Fendt34,35 · Napoleone Ferrara36 · Marcus Fruttiger37 · Dai Fukumura38 · Bart Ghesquière39,40 · Yan Gong19 · Robert J. Grifn29 · Adrian L. Harris41 · Christopher C. W. Hughes12 · Nan W. Hultgren12 · M. Luisa Iruela‑Arispe42 · Melita Irving25 · Rakesh K. Jain38 · Raghu Kalluri43 · Joanna Kalucka20,21 · Robert S. Kerbel44 · Jan Kitajewski45 · Ingeborg Klaassen46 · Hynda K. Kleinmann47 · Pieter Koolwijk48 · Elisabeth Kuczynski44 · Brenda R. Kwak49 · Koen Marien50 · Juan M. Melero‑Martin51 · Lance L. Munn38 · Roberto F. Nicosia5,52 · Agnes Noel53 · Jussi Nurro54 · Anna‑Karin Olsson55 · Tatiana V. Petrova56 · Kristian Pietras57 · Roberto Pili58 · Jefrey W. Pollard59 · Mark J. Post60 · Paul H. A. Quax61 · Gabriel A. Rabinovich62 · Marius Raica23 · Anna M. Randi33 · Domenico Ribatti63,64 · Curzio Ruegg65 · Reinier O. Schlingemann46,48 · Stefan Schulte‑Merker66 · Lois E. H. Smith19 · Jonathan W. Song67,68 · Steven A. Stacker69 · Jimmy Stalin66 · Amber N. Stratman22 · Maureen Van de Velde53 · Victor W. M. van Hinsbergh48 · Peter B. Vermeulen50,72 · Johannes Waltenberger70 · Brant M. Weinstein22 · Hong Xin36 · Bahar Yetkin‑Arik46 · Seppo Yla‑Herttuala54 · Mervin C. -

Report of a Working Group on Vitis: First Meeting
European Cooperative Programme for Plant Genetic Resources Report of a Working ECP GR Group on Vitis First Meeting, 12-14 June 2003, Palić, Serbia and Montenegro E. Maul, J.E. Eiras Dias, H. Kaserer, T. Lacombe, J.M. Ortiz, A. Schneider, L. Maggioni and E. Lipman, compilers IPGRI and INIBAP operate under the name Bioversity International Supported by the CGIAR European Cooperative Programme for Plant Genetic Resources Report of a Working ECP GR Group on Vitis First Meeting, 12-14 June 2003, Palić, Serbia and Montenegro E. Maul, J.E. Eiras Dias, H. Kaserer, T. Lacombe, J.M. Ortiz, A. Schneider, L. Maggioni and E. Lipman, compilers ii REPORT OF A WORKING GROUP ON VITIS: FIRST MEETING Bioversity International is an independent international scientific organization that seeks to improve the well-being of present and future generations of people by enhancing conservation and the deployment of agricultural biodiversity on farms and in forests. It is one of 15 centres supported by the Consultative Group on International Agricultural Research (CGIAR), an association of public and private members who support efforts to mobilize cutting-edge science to reduce hunger and poverty, improve human nutrition and health, and protect the environment. Bioversity has its headquarters in Maccarese, near Rome, Italy, with offices in more than 20 other countries worldwide. The Institute operates through four programmes: Diversity for Livelihoods, Understanding and Managing Biodiversity, Global Partnerships, and Commodities for Livelihoods. The international -

Nation Esa 2015
DIFFE/ RENCES INEQUA/ LITIES AND SOCIO/ LOGICAL IMAGI/ NATION ESA 2015 12TH CONFERENCE OF THE EUROPEAN SOCIOLOGICAL ASSOCIATION 2015 PROGRAMME BOOK DIFFE/ RENCES INEQUA/ LITIES AND SOCIO/ LOGICAL IMAGI/ NATION ESA 2015 12TH CONFERENCE OF THE EUROPEAN SOCIOLOGICAL ASSOCIATION 2015 PROGRAMME BOOK Prague, 25–28 August 2015 ESA 12th Conference Differences, Inequalities and Sociological Imagination www.esa12thconference.eu Organizers IS CAS – Institute of Sociology of the Czech Academy of Sciences – www.soc.cas.cz ESA – European Sociological Association – www.europeansociology.org/ Programme book ISBN 978-80-7330-271-9 PRAGUE, 25–28 AUGUST 2015 ESA 12TH CONFERENCE DIFFERENCES, INEQUALITIES AND SOCIOLOGICAL IMAGINATION PROGRAMME BOOK EUROPEAN SOCIOLOGICAL ASSOCIATION (ESA) INSTITUTE OF SOCIOLOGY OF THE CZECH ACADEMY OF SCIENCES (IS CAS) DIFFERENCES, INEQUALITIES AND SOCIOLOGICAL IMAGINATION DIFFE/ RENCES INEQUA/ LITIES AND SOCIO/ LOGICAL IMAGI/ NATION 6 6 The Theme A profound challenge that the social sciences, and sociology in particular, are now called upon to confront has to do with the depth and extraordinary acceleration of global processes of social and cultural change … … Today's byword 'globalisation' only partially that the international economic crisis has exacerbated captures the full significance of these processes. beyond measure. This situation threatens the very Sociological knowledge therefore encounters existence of democracy and calls for the construction a limitation: it is easier to see what is disappearing of forms of social analysis which are strongly than what is coming into being. Yet this limitation connected to the arena of public policy. Concurrently, can be overturned and become a resource: a stimulus these forms of analysis must also be capable of to intensify our theoretical and empirical exploration offering communities and individuals knowledge and of the world around us by relating everyday life insight that can help to stem the tide of fatalism and to history, connecting individual experiences to apathy. -

Full Catalogue
Catalogue of ELECTRA Papers and SESSION Papers 12/11/2007 Contents The catalogue lists the papers published in ELECTRA after 1967 and the papers discussed at CIGRE Sessions from 1968 on. ELECTRA Papers are listed in chronological order. They are of various nature. The catalogue displays: - the ELECTRA reference - the year of publication - the title of the paper - the nature of the paper - the author(s), either the Working Body which produced it, or the author(s),when it was not the result of a collective work. The Session papers are listed in chronological order also. For each paper are given: - the paper reference, with the year - the title of the paper - the author(s) Structure of the Catalogue -There is a Table of Contents at the beginning of the Catalogue, indexed per year, which helps accessing the right part of the catalogue -The ELECTRA papers follow -The Session papers section comes after. Getting the documents ELECTRA papers and Session papers can be delivered to CIGRE members only. Non members can only get the full sets of Session papers (see the catalogue of Publications) After selection of the papers, members are requested to use the Order Form, available on the Web and send it to the Central Office of CIGRE. This order form gives details on payment, when applying. Documents will be sent in electronic (preferably) or on paper. The service is usually charged for, and charge depends on the nature of the services provided (large number of papers, special request with search…) - Orders will be emailed ([email protected]), posted to: CIGRE (21 rue d’Artois – 75008 Paris) , or faxed (+33 1 53 89 12 99) - Enquiries should be directed to: [email protected], secretary- [email protected], [email protected] (for membership details) IMPORTANT: Intellectual ownership of CIGRE documents. -

Commencement Program 2012, University of Pennsylvania
256 th COMMENCEMENT 256th COMMENCEMENT MAY 14 2012 MAY 14 MAY 2012 UNIVERSITY OF PENNSYLVANIA KEEPING FRANKLIN’S PROMISE In the words of one elegiac tribute, “Great men have two lives: one which occurs while they work on this earth; a second which begins at the day of their death and continues as long as their ideas and conceptions remain powerful.” These words befit the great Benjamin Franklin, whose inventions, innovations, ideas, writings, and public works continue to shape our thinking and renew the Republic he helped to create and the institutions he founded, including the University of Pennsylvania. Nowhere does Franklin feel more contemporary, more revolutionary, and more alive than at the University of Pennsylvania. His startling vision of a secular, nonsectarian Academy that would foster an “Inclination join’d with an Ability to serve Mankind, one’s Country, Friends and Family” has never ceased to challenge Penn to redefine the scope and mission of the modern American university. When pursued vigorously and simultaneously, the two missions – developing the inclination to do good and the ability to do well – merge to help form a more perfect university that educates more capable citizens for our democracy. Penn has embodied and advanced Franklin’s revolutionary vision for 272 years. Throughout its history, Penn has extended the frontiers of higher learning and research to produce graduates and scholars whose work has enriched the nation and all of humanity. The modern liberal arts curriculum as we know it can trace its roots to Franklin’s innovation to have Penn students study international commerce and foreign languages.