Mithramycin Is a Gene-Selective Sp1 Inhibitor That Identifies a Biological Intersection Between Cancer and Neurodegeneration
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Age Dependence of Tumor Genetics in Unfavorable
Cetinkaya et al. BMC Cancer 2013, 13:231 http://www.biomedcentral.com/1471-2407/13/231 RESEARCH ARTICLE Open Access Age dependence of tumor genetics in unfavorable neuroblastoma: arrayCGH profiles of 34 consecutive cases, using a Swedish 25-year neuroblastoma cohort for validation Cihan Cetinkaya1,2, Tommy Martinsson3, Johanna Sandgren1,4, Catarina Träger5, Per Kogner5, Jan Dumanski1, Teresita Díaz de Ståhl1,4† and Fredrik Hedborg1,6*† Abstract Background: Aggressive neuroblastoma remains a significant cause of childhood cancer death despite current intensive multimodal treatment protocols. The purpose of the present work was to characterize the genetic and clinical diversity of such tumors by high resolution arrayCGH profiling. Methods: Based on a 32K BAC whole-genome tiling path array and using 50-250K Affymetrix SNP array platforms for verification, DNA copy number profiles were generated for 34 consecutive high-risk or lethal outcome neuroblastomas. In addition, age and MYCN amplification (MNA) status were retrieved for 112 unfavorable neuroblastomas of the Swedish Childhood Cancer Registry, representing a 25-year neuroblastoma cohort of Sweden, here used for validation of the findings. Statistical tests used were: Fisher’s exact test, Bayes moderated t-test, independent samples t-test, and correlation analysis. Results: MNA or segmental 11q loss (11q-) was found in 28/34 tumors. With two exceptions, these aberrations were mutually exclusive. Children with MNA tumors were diagnosed at significantly younger ages than those with 11q- tumors (mean: 27.4 vs. 69.5 months; p=0.008; n=14/12), and MNA tumors had significantly fewer segmental chromosomal aberrations (mean: 5.5 vs. 12.0; p<0.001). -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Multi-Targeted Mechanisms Underlying the Endothelial Protective Effects of the Diabetic-Safe Sweetener Erythritol
Multi-Targeted Mechanisms Underlying the Endothelial Protective Effects of the Diabetic-Safe Sweetener Erythritol Danie¨lle M. P. H. J. Boesten1*., Alvin Berger2.¤, Peter de Cock3, Hua Dong4, Bruce D. Hammock4, Gertjan J. M. den Hartog1, Aalt Bast1 1 Department of Toxicology, Maastricht University, Maastricht, The Netherlands, 2 Global Food Research, Cargill, Wayzata, Minnesota, United States of America, 3 Cargill RandD Center Europe, Vilvoorde, Belgium, 4 Department of Entomology and UCD Comprehensive Cancer Center, University of California Davis, Davis, California, United States of America Abstract Diabetes is characterized by hyperglycemia and development of vascular pathology. Endothelial cell dysfunction is a starting point for pathogenesis of vascular complications in diabetes. We previously showed the polyol erythritol to be a hydroxyl radical scavenger preventing endothelial cell dysfunction onset in diabetic rats. To unravel mechanisms, other than scavenging of radicals, by which erythritol mediates this protective effect, we evaluated effects of erythritol in endothelial cells exposed to normal (7 mM) and high glucose (30 mM) or diabetic stressors (e.g. SIN-1) using targeted and transcriptomic approaches. This study demonstrates that erythritol (i.e. under non-diabetic conditions) has minimal effects on endothelial cells. However, under hyperglycemic conditions erythritol protected endothelial cells against cell death induced by diabetic stressors (i.e. high glucose and peroxynitrite). Also a number of harmful effects caused by high glucose, e.g. increased nitric oxide release, are reversed. Additionally, total transcriptome analysis indicated that biological processes which are differentially regulated due to high glucose are corrected by erythritol. We conclude that erythritol protects endothelial cells during high glucose conditions via effects on multiple targets. -

Literature Mining Sustains and Enhances Knowledge Discovery from Omic Studies
LITERATURE MINING SUSTAINS AND ENHANCES KNOWLEDGE DISCOVERY FROM OMIC STUDIES by Rick Matthew Jordan B.S. Biology, University of Pittsburgh, 1996 M.S. Molecular Biology/Biotechnology, East Carolina University, 2001 M.S. Biomedical Informatics, University of Pittsburgh, 2005 Submitted to the Graduate Faculty of School of Medicine in partial fulfillment of the requirements for the degree of Doctor of Philosophy University of Pittsburgh 2016 UNIVERSITY OF PITTSBURGH SCHOOL OF MEDICINE This dissertation was presented by Rick Matthew Jordan It was defended on December 2, 2015 and approved by Shyam Visweswaran, M.D., Ph.D., Associate Professor Rebecca Jacobson, M.D., M.S., Professor Songjian Lu, Ph.D., Assistant Professor Dissertation Advisor: Vanathi Gopalakrishnan, Ph.D., Associate Professor ii Copyright © by Rick Matthew Jordan 2016 iii LITERATURE MINING SUSTAINS AND ENHANCES KNOWLEDGE DISCOVERY FROM OMIC STUDIES Rick Matthew Jordan, M.S. University of Pittsburgh, 2016 Genomic, proteomic and other experimentally generated data from studies of biological systems aiming to discover disease biomarkers are currently analyzed without sufficient supporting evidence from the literature due to complexities associated with automated processing. Extracting prior knowledge about markers associated with biological sample types and disease states from the literature is tedious, and little research has been performed to understand how to use this knowledge to inform the generation of classification models from ‘omic’ data. Using pathway analysis methods to better understand the underlying biology of complex diseases such as breast and lung cancers is state-of-the-art. However, the problem of how to combine literature- mining evidence with pathway analysis evidence is an open problem in biomedical informatics research. -

Fanyue Sun-Dissertation.Pdf
Towards the identification of sex-determining gene(s) in channel catfish by Fanyue Sun A dissertation submitted to the Graduate Faculty of Auburn University in partial fulfillment of the requirements for the Degree of Doctor of Philosophy Auburn, Alabama August 3rd, 2013 Keywords: sex determination, sex chromosome, next-generation sequencing, genome assembly and scaffolding, comparative analysis, catfish Copyright 2013 by Fanyue Sun Approved by Zhanjiang Liu, Chair, Alumni Professor of Fisheries and Allied Aquacultures Joanna Wysocka-Diller, Associate Professor of Biological Sciences Nannan Liu, Professor of Plant Pathology and Entomology Rex Dunham, Alumni Professor of Fisheries and Allied Aquacultures Abstract Sex is the most fundamental feature in the life of an organism. Studying of sex determination is an important area in animal developmental and evolutionary biology, as well as ecology. In teleost, sex determination mechanism exhibits extraordinary plasticity and diversity with respect to the evolutionary pattern. Catfish have a XY male/XX female sex chromosome system. The exact mechanism of sex determination in catfish is unknown at present. As a first step towards the identification of sex-determining genes in catfish, we performed the first transcriptome-level analysis of the catfish testis using high throughput Illumina sequencing to understand the transcriptome of the catfish testes. Gene ontology and annotation analysis suggested that many of the male-biased genes identified from the analysis were involved in gonadogenesis, spermatogenesis, testicular determination, gametogenesis, gonad differentiation, and possibly sex determination. Our analysis would lay the basis for further follow-up analysis of genes involved in sex determination and differentiation in catfish. To move toward the goal of identification of the Y-specific fragments in channel catfish, we utilized multiple approaches to get the best assembly and scaffolding of X and Y chromosome in catfish and conducted in silico comparative analysis between these two chromosomes. -

(A) Schematic Representation of the Secondary Assays Done to Control for Promoter Or Luciferase Specific Effects
Supplementary Figure 1. Secondary Assays (A) Schematic representation of the secondary assays done to control for promoter or luciferase specific effects. The Hh-GL2 Assay is the original assay format. The Hh-GL3 assay is similar to the original reporter construct, but lacks a 3' intron that contributed to nonsense mediated decay of the reporter (see Materials and Methods). In the luciferase reversal assay, the promoters and luciferase constructs were swapped. In the promoter matching assay, the Hh expressing construct was driven by a Pol III promoter. (B) Ci overexpression partially activates the Hh signaling pathway and can partially rescue loss of reporter activity due to RNAi elimination of smo. Graphic representation of normalized luciferase values for the indicated stimulus (either Hh or Ci, in equal amounts) and dsRNA treatment. Ci expression activates the ptc∆136 reporter to about half the level induced by Hh treatment. smo dsRNA almost completely eliminates reporter activity when Hh is used as stimulus, while smo dsRNA only reduces reporter by about 30% when Ci is used as stimulus. Supplementary Figure 1 A B Hh Ci Ptc∆136 Firefly GL2 Assay 70 Pol 3 Renilla 60 50 Actin Hedgehog FL 40 30 20 GL3 10 Ptc∆136 Firefly Assay 0 Normalized Value Pol 3 Renilla GFP Smo 5' Ci 5' Actin Hedgehog FL dsRNA Luciferase Pol 3 Firefly Reversal Assay Ptc∆136 Renilla Actin Hedgehog FL Promoter Ptc∆136 Firefly Matching Pol 3 Renilla Assay Pol 3 Hedgehog FL Supplementary Figure 2. Phosphorylation of Fu and Cos2 are not affected by reduction in Cdk9, Pitslre, or mts. -
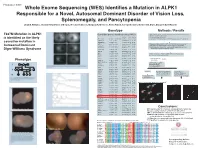
T237M Mutation in ALPK1 Is Identified As the Likely Causative Mutation In
Program #: 3367 Whole Exome Sequencing (WES) Identifies a Mutation in ALPK1 Responsible for a Novel, Autosomal Dominant Disorder of Vision Loss, Splenomegaly, and Pancytopenia Lloyd B. Williams, Chad D. Huff, Denise J. Morgan, Rosann Robinson, Margaux A. Morrison, Krista Kinard, George Rodgers, Kathleen B. Digre, Margaret M. DeAngelis Genotype Methods / Results WES on 4 individuals - 2 affected II-3 III-2, and 2 unaffected II-2 and III-3 T237M Mutation in ALPK1 Candi date genes identified using VAAST Illuimina Truseq Enrichment Kit position Allele Protein Quantitative PCR enrichment is identified as the likely Gene name p-value chromosome (hg19) change change Sequencing with Illumina HiSeq 2000 101 cycle paired end sequencing PRAMEF11 6.10E-06 chr1 12887174 C->T R->H causative mutation in ANKRD20A4 7.32E-06 chr9 69423637 G->A E->K MRPL4 8.55E-06 chr19 10367459 C->T R->W Mapped and aligned with Picard Tools (http://picard.sourceforge.net) FAM90A10 9.77E-06 chr8 7629232 G->T A->S SNPs and indels identified with Genome analysis toolkit (GATK) Autosomal Dominant Manual inspection and curation was done with Intergrative Genomics GOLGA6L10 9.77E-06 chr15 82635194 T->C E->G Viewer (http://www.broadinstitute.org/igv) Digre-Williams Syndrome FAM90A20 1.28E-05 chr8 7155458 C->G A->G EEF1A1 1.65E-05 chr6 74228474 C->T R->H MSI1 1.65E-05 chr12 120800875 C->T V->M VAAST - compares to HGMD, dbSNP, ESP, and1000 Genomes VDAC2 1.71E-05 chr10 76980685 G->T A->S panels to rule out common variants. PIM1 2.08E-05 chr6 37138779 A->T K->M USP11 2.26E-05 chrX -

Systematic Analysis of the Transcriptome Profiles and Co-Expression Networks of Tumour Endothelial Cells Identifies Several Tumo
cancers Article Systematic Analysis of the Transcriptome Profiles and Co-Expression Networks of Tumour Endothelial Cells Identifies Several Tumour-Associated Modules and Potential Therapeutic Targets in Hepatocellular Carcinoma Thomas Mohr 1,2 , Sonja Katz 1 , Verena Paulitschke 3 , Nadim Aizarani 4 and Alexander Tolios 5,6,7,∗ 1 ScienceConsult—DI Thomas Mohr KG, Enzianweg 10a, A-2353 Guntramsdorf, Austria; [email protected] or [email protected] (T.M.); [email protected] (S.K.) 2 Institute of Cancer Research, Department of Medicine I, Medical University of Vienna and Comprehensive Cancer Center, A-1090 Vienna, Austria 3 Department of Dermatology, Medical University of Vienna, A-1090 Vienna, Austria; [email protected] 4 Max-Planck-Institute of Immunobiology and Epigenetics, D-79108 Freiburg, Germany; [email protected] 5 Department of Blood Group Serology and Transfusion Medicine, Medical University of Vienna, A-1090 Vienna, Austria 6 Center of Physiology and Pharmacology, Institute of Vascular Biology and Thrombosis Research, Medical University of Vienna, A-1090 Vienna, Austria 7 Section of Artificial Intelligence and Decision Support, Center for Medical Statistics, Informatics and Citation: Mohr, T.; Katz, S.; Intelligent Systems, Medical University of Vienna, A-1090 Vienna, Austria Paulitschke, V.; Aizarani, N.; Tolios, * Correspondence: [email protected] A. Systematic Analysis of the Transcriptome Profiles and Simple Summary: Endothelial cells, the innermost layer of blood vessels, play an essential role in the Co-Expression Networks of Tumour progression of cancer, particularly liver cancer. To develop cancer therapies targeting those cells, the Endothelial Cells Identifies Several investigation of gene co-expression networks is of great importance. -
Protein Network Analysis Indicates That Ebola Virus, Neisseria Meningitidis and Trypanosoma Brucei Trigger Common Host Defense Response Pathways
Protein network analysis indicates that Ebola virus, Neisseria meningitidis and Trypanosoma brucei trigger common host defense response pathways Shishir K Gupta Julius-Maximilians-Universitat Wurzburg Alicia Ponte-Sucre Universidad Central de Venezuela Instituto de Biomedicina Elena Bencurova Julius-Maximilians-Universitat Wurzburg Thomas Dandekar ( [email protected] ) University of Würzburg https://orcid.org/0000-0003-1886-7625 Research article Keywords: blood-borne pathogen; interactome; protein-protein interaction; life cycle; hub proteins; antibiotic targeting; pathway analysis; Ebola virus; Neisseria meningitidis; Human African Trypanosomiasis Posted Date: October 21st, 2019 DOI: https://doi.org/10.21203/rs.2.16270/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Version of Record: A version of this preprint was published at Computational and Structural Biotechnology Journal on September 16th, 2021. See the published version at https://doi.org/10.1016/j.csbj.2021.09.017. Page 1/25 Abstract Background: Sepsis is the most severe nal state of infection. Might pathogens belonging to different kingdoms of life and pathogen classes trigger analogous responses pathways in the host? Is there a common denominator or strategy to prevent it? Here we use interactomics and comparative evolutionary analysis of three serious infectious pathogens, spanning viruses, bacteria and parasites: Filovirus ebolavirus (ZE; Zaire ebolavirus , Bundibugyo ebolavirus), Neisseria meningitides (NM), and Trypanosoma brucei (Tb) that target the blood during their infectious life cycle. Results: We analyze 2797 unique human proteins targeted by the pathogens. The comparison resulted in specic and shared protein-protein interactions (PPIs) with each pathogen, derived from orthology searches of experimentally validated PPIs. -

Distinct Transcriptomes Define Rostral and Caudal 5Ht Neurons
DISTINCT TRANSCRIPTOMES DEFINE ROSTRAL AND CAUDAL 5HT NEURONS by CHRISTI JANE WYLIE Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy Dissertation Advisor: Dr. Evan S. Deneris Department of Neurosciences CASE WESTERN RESERVE UNIVERSITY May, 2010 CASE WESTERN RESERVE UNIVERSITY SCHOOL OF GRADUATE STUDIES We hereby approve the thesis/dissertation of ______________________________________________________ candidate for the ________________________________degree *. (signed)_______________________________________________ (chair of the committee) ________________________________________________ ________________________________________________ ________________________________________________ ________________________________________________ ________________________________________________ (date) _______________________ *We also certify that written approval has been obtained for any proprietary material contained therein. TABLE OF CONTENTS TABLE OF CONTENTS ....................................................................................... iii LIST OF TABLES AND FIGURES ........................................................................ v ABSTRACT ..........................................................................................................vii CHAPTER 1 INTRODUCTION ............................................................................................... 1 I. Serotonin (5-hydroxytryptamine, 5HT) ....................................................... 1 A. Discovery.............................................................................................. -

Use Tumor Suppressor Genes As Biomarkers for Diagnosis Of
www.nature.com/scientificreports OPEN Use tumor suppressor genes as biomarkers for diagnosis of non‑small cell lung cancer Chuantao Zhang1,3, Man Jiang1,3, Na Zhou1, Helei Hou1, Tianjun Li1, Hongsheng Yu1, Yuan‑De Tan2* & Xiaochun Zhang1* Lung cancer is the leading cause of death worldwide. Especially, non‑small cell lung cancer (NSCLC) has higher mortality rate than the other cancers. The high mortality rate is partially due to lack of efcient biomarkers for detection, diagnosis and prognosis. To fnd high efcient biomarkers for clinical diagnosis of NSCLC patients, we used gene diferential expression and gene ontology (GO) to defne a set of 26 tumor suppressor (TS) genes. The 26 TS genes were down‑expressed in tumor samples in cohorts GSE18842, GSE40419, and GSE21933 and at stages 2 and 3 in GSE19804, and 15 TS genes were signifcantly down‑expressed in tumor samples of stage 1. We used S‑scores and N‑scores defned in correlation networks to evaluate positive and negative infuences of these 26 TS genes on expression of other functional genes in the four independent cohorts and found that SASH1, STARD13, CBFA2T3 and RECK were strong TS genes that have strong accordant/discordant efects and network efects globally impacting the other genes in expression and hence can be used as specifc biomarkers for diagnosis of NSCLC cancer. Weak TS genes EXT1, PTCH1, KLK10 and APC that are associated with a few genes in function or work in a special pathway were not detected to be diferentially expressed and had very small S‑scores and N‑scores in all collected datasets and can be used as sensitive biomarkers for diagnosis of early cancer. -
SUPPLEMENTARY APPENDIX Post-Transplant Molecularly Defined Burkitt Lymphomas Are Frequently MYC -Negative and Characterized by the 11Q-Gain/Loss Pattern
SUPPLEMENTARY APPENDIX Post-transplant molecularly defined Burkitt lymphomas are frequently MYC -negative and characterized by the 11q-gain/loss pattern Julio Finalet Ferreiro, 1 Julie Morscio, 2 Daan Dierickx, 3 Lukas Marcelis, 2 Gregor Verhoef, 3 Peter Vandenberghe, 1 Thomas Tousseyn, 2 and Iwona Wlodarska 1 1KU Leuven, University of Leuven, Center for Human Genetics; 2KU Leuven, University of Leuven, Translational Cell and Tissue Research and KU Leuven, University Hospitals Leuven, Department of Pathology; and 3KU Leuven, University Hospitals Leuven, Department of Hematology, Belgium Supplementary information Supplementary Methods Cytogenetics and FISH G-banding chromosomal analysis and fluorescence in situ hybridization (FISH) followed routine methods. DNA probes, Bacterial Artificial Clones (BAC), applied for FISH were selected from www.Oct29012.archive.ensembl.org (Supplementary Figure 2). The probes were labeled with SpectrumOrange- and SpectrumGreen-d-UTP (Abbott Molecular, Ottigne, Belgium) using random priming. FISH images were acquired with a fluorescence microscope equipped with an Axiophot 2 camera (Carl Zeiss Microscopy, Jena, Germany) and a MetaSystems ISIS imaging system (MetaSystems, Altlussheim, Germany). In each experiment up to 8 abnormal metaphases and/or 200 interphase cells were evaluated. High resolution array Comparative Genome Hybridization (aCGH) Total genomic DNA was isolated from fresh frozen lymphoma samples using standard procedures. Genomic profiling, following the manufacturer’s protocols, was performed using the Affymetrix Cytogenetic array 2.7M (www.affymetrix.com). The initial data analysis was performed with the software “Chromosome Analysis Suit” (CHAS) and subsequent analysis (segmentation and aberration heatmap) were performed using the software “ArrayStudio” (www.omicsoft.com). Array CGH data are available at GEO (http://www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc=GSE64086).